Primer set for detecting American canine influenza virus subtype H3N8 and application thereof
A technology of canine influenza virus, H3N8, which is applied to the detection of North American H3N8 subtype canine influenza virus primer set and its application field, which can solve the problems of easy mutual influence, cumbersome operation, and low sensitivity, and achieve great application value and simple operation , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1, the design of primer set
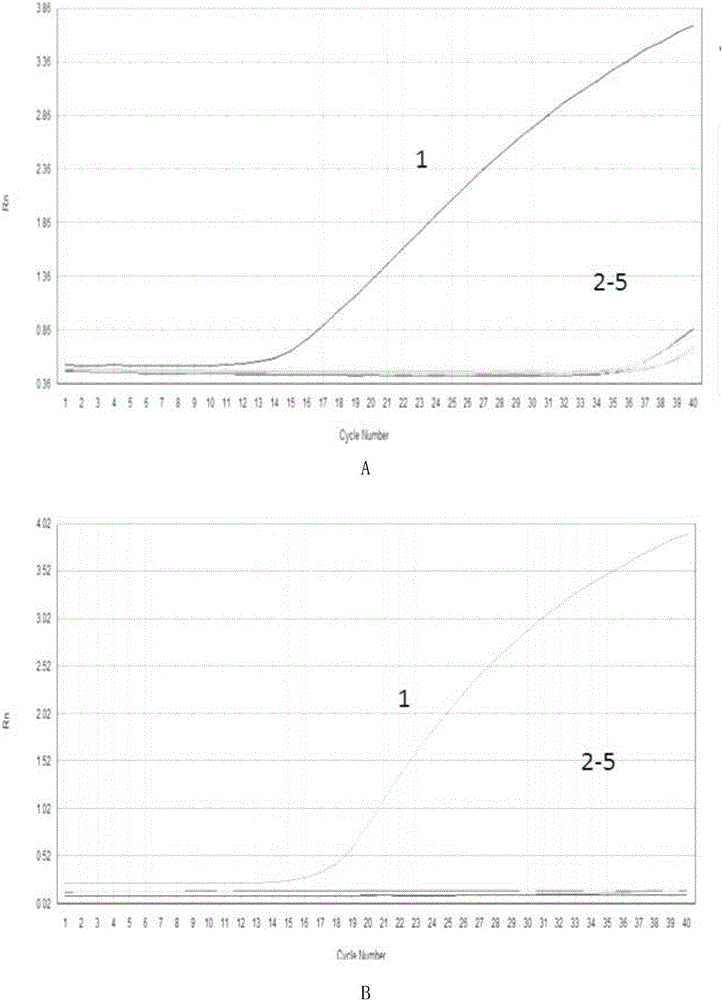
[0043] After a large number of sequence analysis, multiple primer pairs were obtained, and the specificity and sensitivity of each primer pair were tested through preliminary experiments, and finally two primer pairs for identifying H3N8 subtype canine influenza virus were screened out.
[0044] Primer pair A (the target sequence is located in the HA gene, the annealing temperature is 59° C.) consists of primer F1 and primer R1.
[0045] Primer pair B (the target sequence is located in the NA gene, and the annealing temperature is 58° C.) consists of primer F2 and primer R2.
[0046] Primer F1 (sequence 1 of the sequence listing): 5'-TTGCTACCCATATGACATCCCTGAC-3';
[0047] Primer R1 (SEQ ID NO: 2 of the Sequence Listing): 5'-GTGAATCCCTCTGCTGTGAATTCCA-3'.
[0048] Primer F2 (sequence 3 of the sequence listing): 5'-CCCCAATGAAGGGAAGGTGGAATGC-3;
[0049] Primer R2 (SEQ ID NO: 4 of the Sequence Listing): 5'-ATCCTCTCCCCTAGGGGTGTCAGTG...
Embodiment 2
[0050] Embodiment 2, establishment of method
[0051] 1. Extract the total RNA of the sample to be tested and reverse transcribe it into cDNA.
[0052] The sample to be tested can be a tissue to be tested or a virus to be tested. The tissue to be tested can be canine liver tissue, canine brain tissue, canine lung tissue, canine nasal cavity swab, etc.
[0053] 2. Use the cDNA obtained in step 1 as a template, and use the primers designed in Example 1 to perform fluorescent quantitative PCR on A, and monitor the fluorescence intensity in real time.
[0054] Fluorescent quantitative PCR reaction system (20 μL): SYBR Premix Ex Taq (2X) 10 μL, primer F1 0.5 μL, primer R1 0.5 μL, cDNA 2 μL, add double distilled water to 20 μl.
[0055] The reaction program of fluorescent quantitative PCR: 95°C for 10min; 95°C for 15s, 59°C for 1min, 40 cycles.
[0056] 3. Using the cDNA obtained in step 1 as a template, use the primers designed in Example 1 to carry out fluorescent quantitative ...
Embodiment 3
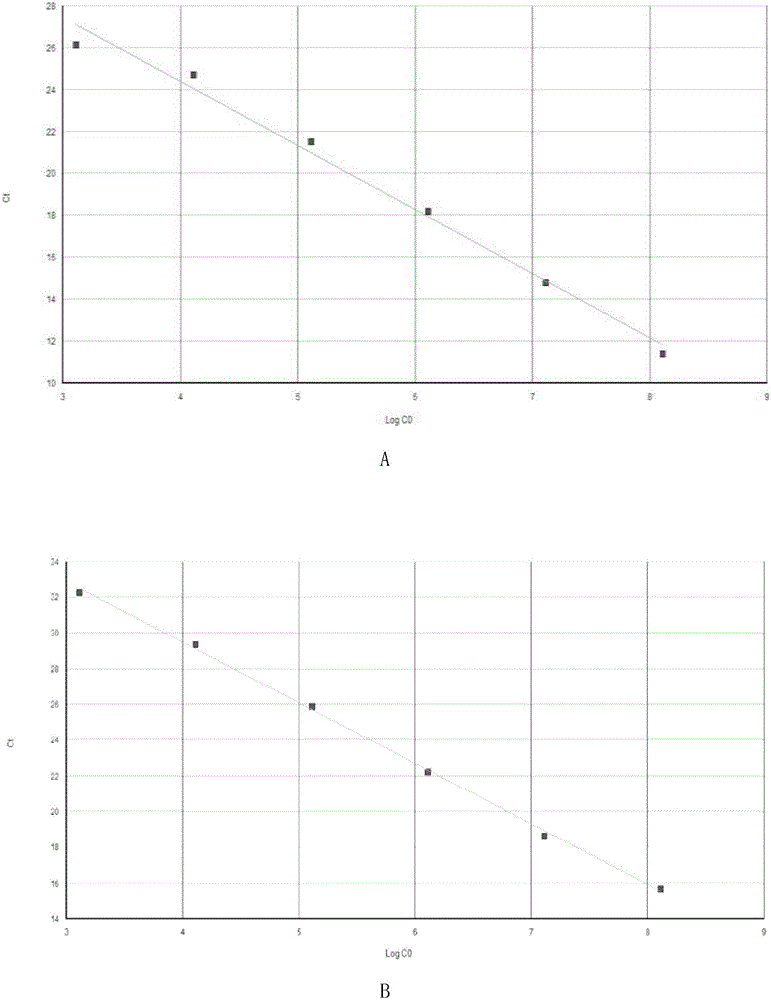
[0062] Embodiment 3, establishment of standard curve
[0063] Gene template copy number = DNA mass concentration / DNA molecular weight.
[0064] 1. Preparation of standard quality grain A
[0065] The DNA molecule shown in sequence 5 in the sequence listing is the HA gene fragment of North American H3N8 subtype canine influenza virus. Insert the DNA molecule shown in sequence 5 in the sequence listing into the multiple cloning site of pUC57 vector to obtain standard plasmid A.
[0066] 2. Preparation of standard plasmid B
[0067] The DNA molecule shown in sequence 6 in the sequence listing is the NA gene fragment of North American H3N8 subtype canine influenza virus. The DNA molecule shown in sequence 6 of the sequence listing was inserted into the multiple cloning site of the pUC57 vector to obtain standard plasmid B.
[0068] 3. Establishment of standard curve A
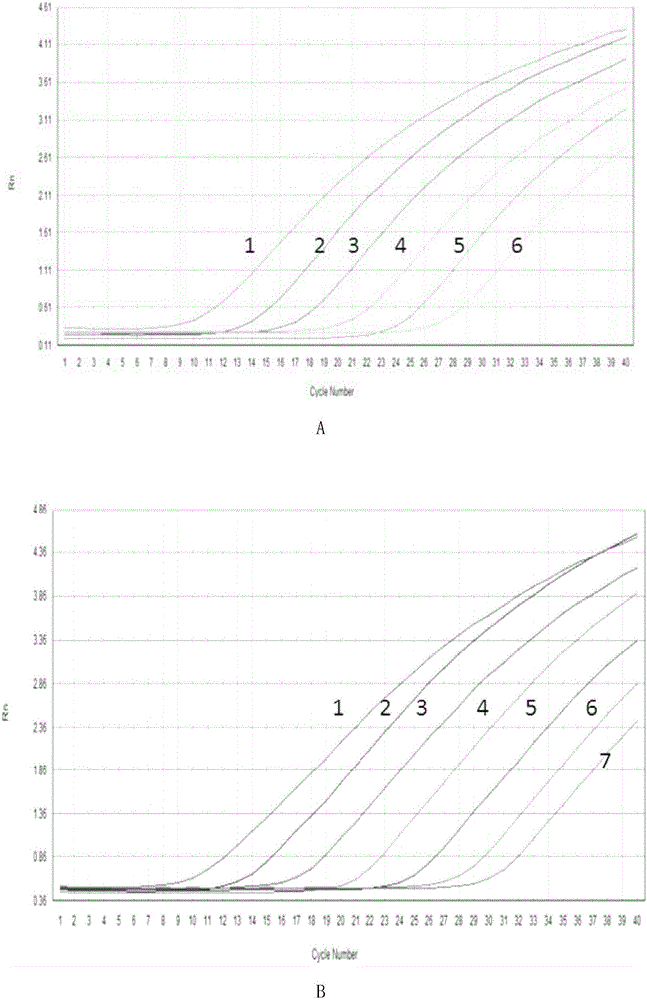
[0069] 1. Gradiently dilute the standard quality Granules with double distilled water 10 times to obtain a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com