Determining cancer agressiveness, prognosis and responsiveness to treatment
An invasive, cancer-based technology for specific peptides, chemical instruments and methods, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0386] Materials and methods
[0387] Meta-analysis of global gene expression in TNBC
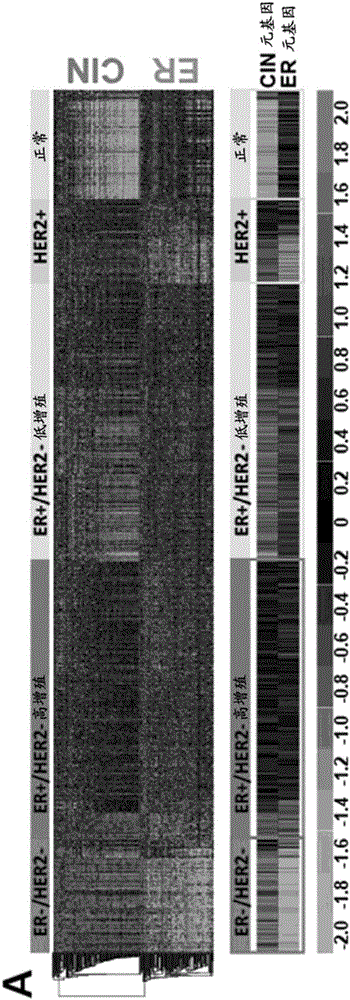
[0388] In Oncomine TM database 19 (Compendia Bioscience, MI) using breast cancer primary filter (130 datasets), sample filter to use clinical specimens and dataset filter to use mRNA dataset with more than 151 patients (22 datasets) , we performed a global gene expression meta-analysis. Patients of all ages, sexes, disease stages, or treatments were included. Three additional filters were applied for three independent differential analyses: (1) triple negative (TNBC cases vs non-TNBC cases, 8 data sets 49-56 ); (2) Metastasis event analysis at 5 years (transfer event vs non-transfer event, 7 data sets 53,54,57-61 ) and (3) survival at 5 years (patients who died vs. patients who survived, 7 datasets 49,54,56,58,61-63 ). Dysregulated genes were selected based on the median p-value of the median gene rank in overexpression pattern or underexpression pattern in the dataset ( Figure 8 )...
Embodiment 2
[0545] Materials and methods
[0546] Meta-analysis of global gene expression in TNBC
[0547] In Oncomine TM Database [37] (Compendia Bioscience, Ann Arbor, MI) using primary filter for breast cancer (130 datasets), sample filter to use clinical specimens and dataset filter to use mRNA data with more than 151 patients set (22 datasets), we performed a meta-analysis of global gene expression. Two additional filters were used to perform two independent differential analyses. The first differential analysis was an analysis of metastatic events at 5 years (metastatic events vs. patients, 7 datasets [39, 57, 59, 61-64]). For each of the two differential analyses, dysregulated genes were selected based on the median p-value of the median gene rank for either the overexpression pattern or the underexpression pattern in the dataset.
[0548] Export 28-imprint (TN imprint)
[0549] The online tool KM-Plotter [38], which collates gene expression data of more than 4000 breast canc...
Embodiment 3
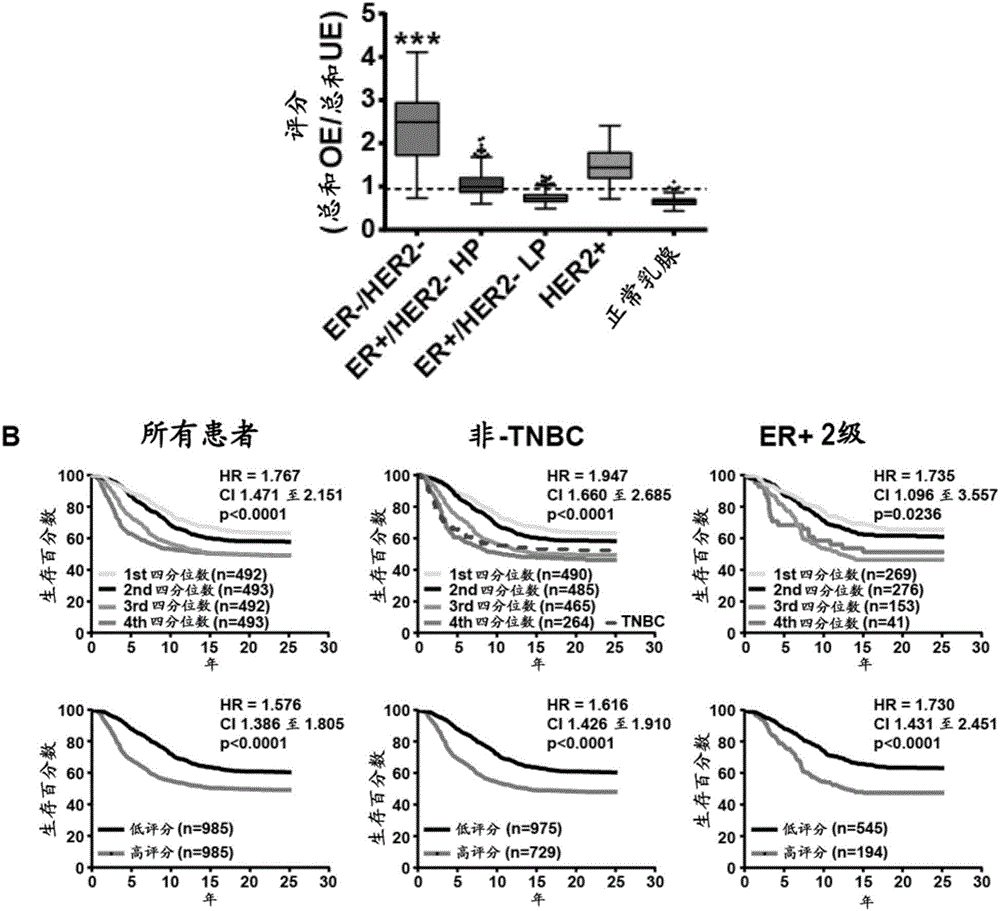
[0751] The iBCR test presented herein was developed from a meta-analysis of gene expression profiles of breast cancer. The test is based on the expression of 43 genes that are predictive as signatures in breast cancer (regardless of subtype). The test was also found to be predictive in lung adenocarcinoma. Patients with high iBCR scores had much worse overall survival than those with low iBCR scores.
[0752] In the current study, the Cancer Genome Atlas (TCGA) dataset was investigated for multiple cancer types for three purposes. First, to determine the differences at the protein level between breast cancer cases with high iBCR scores and breast cancer cases with low iBCR scores. This comparison was also performed for lung adenocarcinoma. Second, to determine whether dysregulated proteins / phosphoproteins between high and low iBCR score tumors are predictive. Finally, the prognostic value of iBCR miRNA signature and associated protein signature was predictive in other canc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com