Method for performing forest tree multiple-trait pyramiding breeding based on multiple-trait genomic selection
A genome selection and aggregation breeding technology, applied in the field of forest tree genetics and breeding, can solve the problems of long required period and inability to accurately select parents, and achieve the effect of reducing workload, shortening breeding period, reliable productivity and genetic background.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The forest tree multi-character aggregation breeding method of the present invention comprises the steps of collection of clone individual phenotype data, clone pedigree information and A matrix construction, SNP marker typing data and G matrix construction, multi-character model establishment and data analysis steps; specifically as follows:
[0048] 1. Clonal individual phenotype data collection
[0049] (1) The test material is: Eucalyptus clone test forest, 165 test clones in total, about 40 plants for each clone.
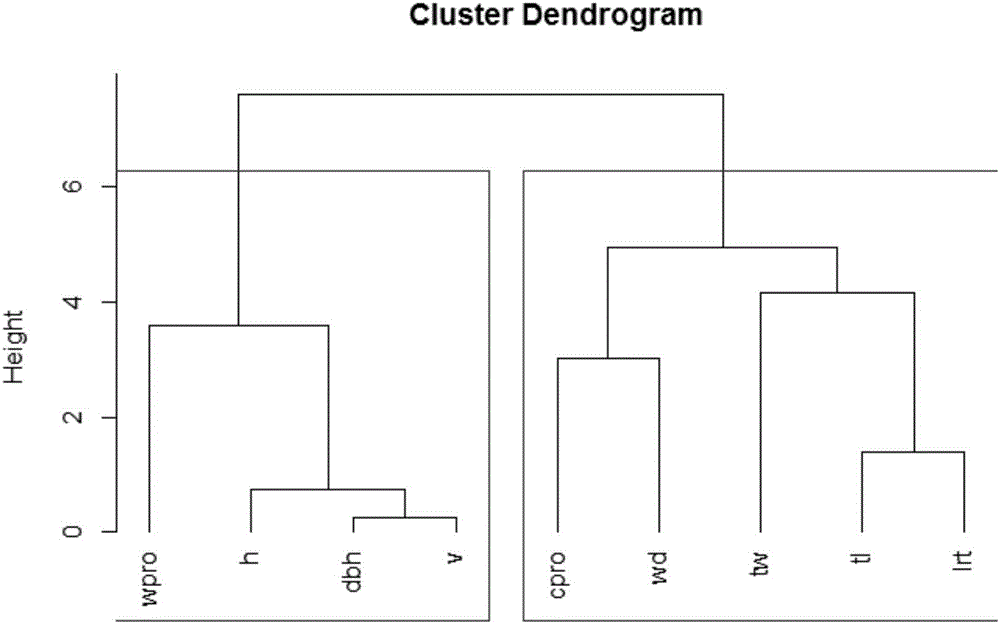
[0050] Sampling 30 eucalyptus clones, 5 plants for each clone, measured tree height (h), diameter at breast height (dbh), volume (v), heartwood ratio (cpro), wood density (wd), wood water absorption (wpro) , tracheid length (tl), tracheid width (tw) and tracheid length-to-width ratio (lrt), and then perform cluster analysis of traits, classify according to positive and negative correlations, and obtain two major classification information of traits;
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com