Flow correction method for second-generation cancer genome high-throughput sequencing data
A technology for sequencing data and correction methods, applied in the field of data science, can solve problems such as inapplicable tumor genome data processing procedures, and achieve the effects of saving storage space, improving accuracy, and improving computing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
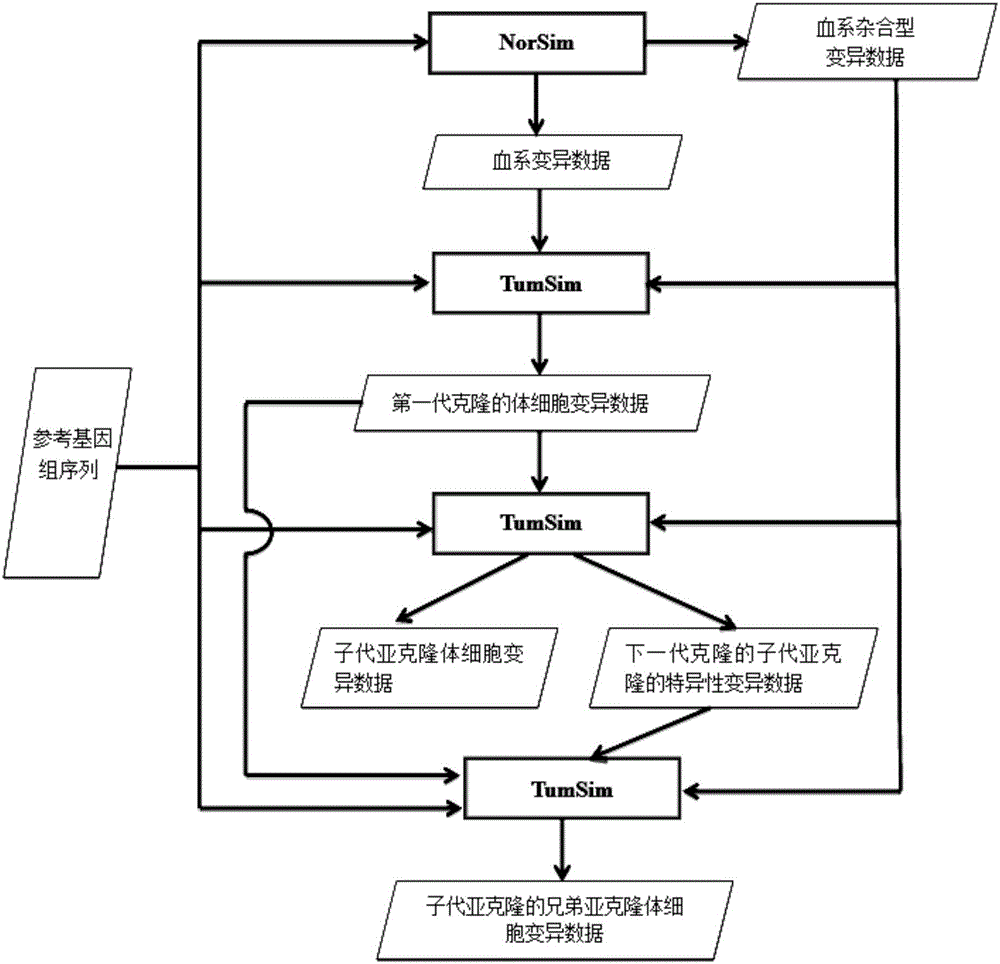
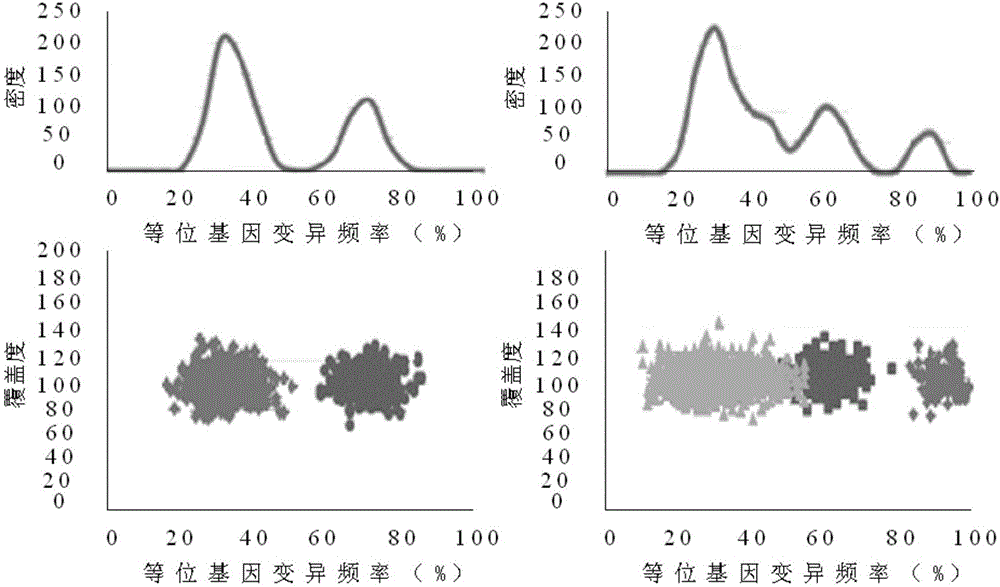
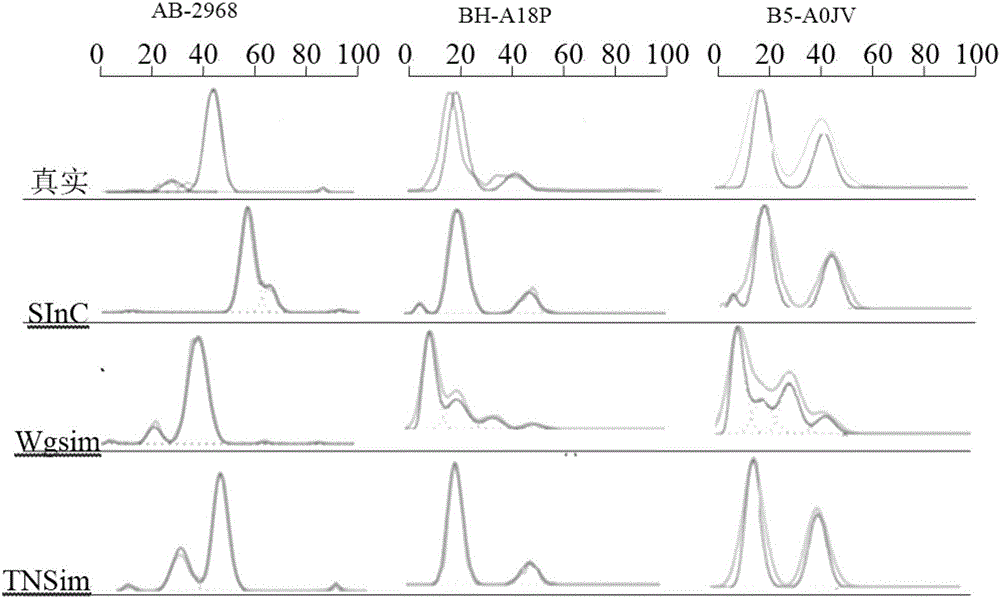
[0064] see figure 1 As shown, the present invention discloses a process correction method for high-throughput sequencing data of the second-generation tumor genome. A 32-bit unsigned number is used as the identification quantity to record the variation data of each blood line, and the generated data reflects the purity and different subgroups. According to the read segment data of the clone ratio, the somatic variation calibration data of the brother subclones of the offspring subclones are obtained according to the bloodline variation data relationship of father-son subclones inheritance and brother subclones mutually exclusive relationship, and are used for the second The processing flow of high-throughput sequencing data of generation tumor genome was corrected.
[0065] Among them, the mutation data algorithm of father-son subclone inheritance and sibling subclone mutual exclusion relationship is as follows:
[0066] S1. As mentioned above, tumor cells inherit bloodline v...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com