Kit for rapidly detecting SLCO1B1 gene mutation and detection method
A detection method and kit technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of poor specificity and low detection accuracy, and achieve the effect of easy operation, simple equipment requirements, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] (1), extract blood DNA, and dilute to 100ng / ul;
[0042] (2) Take 20ul ARMS-PCR reaction system, specifically including: 10×PCR Buffer 2μl, FI 1μl, RI 1μl, FO 5μl, RO 5μl dNTP mixture 1.6μl, Taq DNA polymerase 0.4μl, Mg2+1.6μl, DNA Template 1μl, add ddH2O to 20μl;
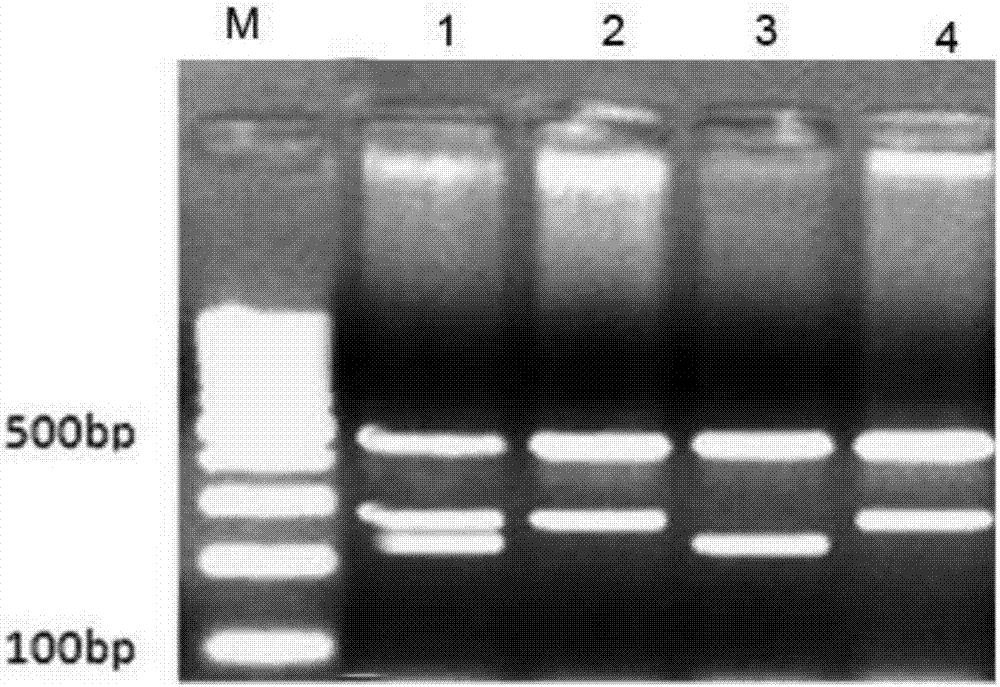
[0043] (3) Put the mixed reaction solution into the PCR instrument, set the program as follows: pre-denaturation at 95°C for 10 minutes, then enter 1 cycle, denaturation at 95°C for 15s, annealing at 45°C for 45s, extension at 72°C for 45s, a total of 32 cycles Cycle, and then continue to extend at 72° C. for 8 min, 1 cycle, and then analyze the PCR reaction product by gel electrophoresis using 3% agarose gel for gel electrophoresis analysis.
[0044] The present invention initially designed 4 sets of primers for primer screening, the specific sequences are as follows:
[0045] FI-1: 5'-TCTTATCTACATAGGTTGTTTAAAGGATTC-3'
[0046] RI-1:5'-ACCATATATCCACATGTATGACCGAC-3'
[0047] FO-1: 5'-CAGTCTCAGGTATGTATTTA...
Embodiment 2
[0063] (1), extract blood DNA, and dilute to 100ng / ul;
[0064] (2) Take 20ul ARMS-PCR reaction system, specifically including: 10×PCR Buffer 2μl, FI 1μl, RI 1μl, FO 5μl, RO 5μl dNTP mixture 1.6μl, Taq DNA polymerase 0.4μl, Mg2+1.6μl, DNA Template 1μl, add ddH2O to 20μl;
[0065] (3) Put the mixed reaction solution into the PCR instrument, set the program as follows: pre-denaturation at 95°C for 10 minutes, then enter 1 cycle, denaturation at 95°C for 15s, annealing at 45°C for 45s, extension at 72°C for 45s, a total of 32 cycles Cycle, and then continue to extend at 72° C. for 8 min, 1 cycle, and then analyze the PCR reaction product by gel electrophoresis using 3% agarose gel for gel electrophoresis analysis.
[0066] PCR reaction products were analyzed by gel electrophoresis using 3% agarose gel.
[0067] The ratio of endogenous primer and exogenous primer in embodiment 2 is 1:1.
Embodiment 3
[0069] (1), extract blood DNA, and dilute to 100ng / ul;
[0070] (2) The optimal reaction system for ARMS-PCR is 20ul, specifically including: 10×PCR Buffer 2μl, FI 1μl, RI 1μl, FO 10μl, RO 10μl dNTP mixture 1.6μl, Taq DNA polymerase 0.4μl, Mg2+1.6μl, DNA template 1μl, add ddH2O to 20μl;
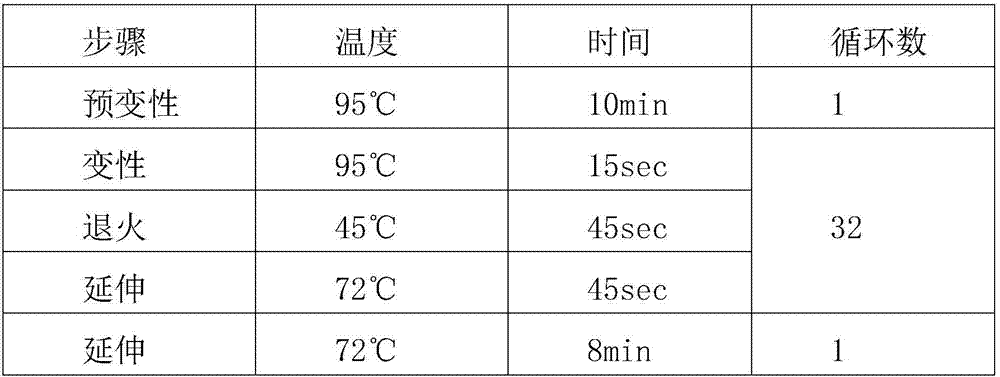
[0071] (3), put the mixed reaction solution into the PCR instrument, the setting procedure is as follows:
[0072]
[0073] PCR reaction products were analyzed by gel electrophoresis using 3% agarose gel.
[0074] The ratio of endogenous primer and exogenous primer in embodiment 2 and 3 is respectively 1:1 and 1:10, all do not obtain ideal result, so applicable endogenous primer and exogenous primer ratio in the present invention are 1:5 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com