Drug target prediction and drug comprehensive evaluation method
An evaluation method and drug technology, applied in biochemical equipment and methods, drug combinations, pharmaceutical formulations, etc., can solve the problem that the coverage of evaluation indicators is difficult to achieve ideal results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
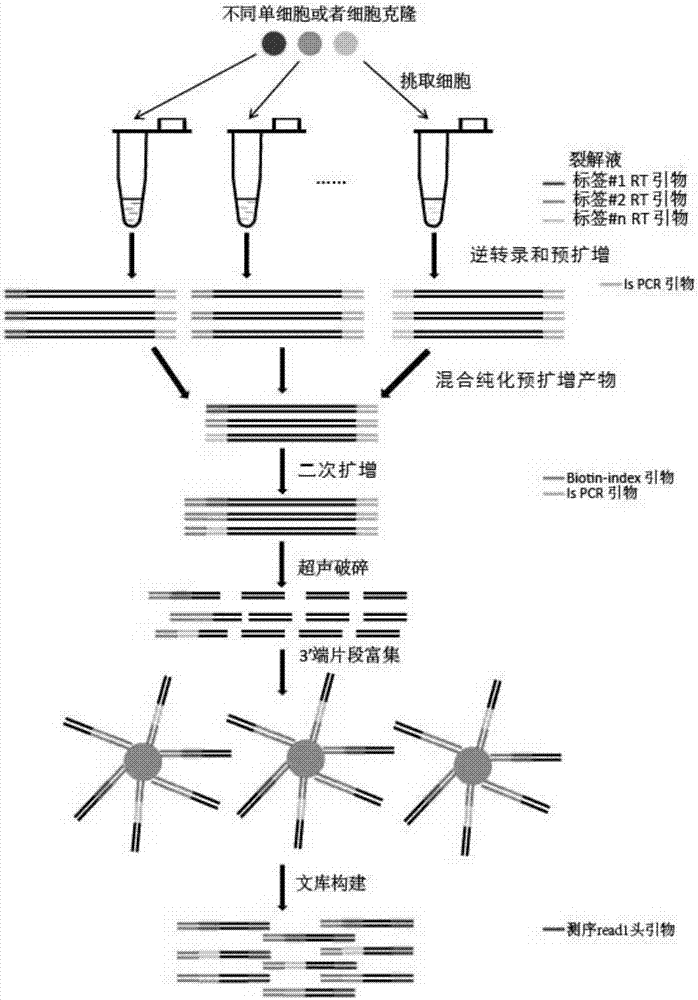
[0075] Example 1. Construction of a multi-label whole transcriptome sequencing library
[0076] (1) Cell culture
[0077] 1. Culture and passage of HepG2 liver cancer cell line
[0078] (1) The HepG2 liver cancer cell line was cultured in DMEM-HG medium containing 10% FBS to a density close to 80%.
[0079] (2) Wash twice with PBS buffer to remove dead cells on the upper layer, add an appropriate amount of Trypsin to digest at 37°C for 3 minutes, then neutralize with DMEM-HG medium with 10% FBS, centrifuge and remove the supernatant.
example 1
[0080] (3) Resuspend in DMEM-HG medium with 10% FBS, count and pass, the passage ratio is 1:6.
[0081] 2. Test the effectiveness of FDA-approved targeted drugs on the HepG2 liver cancer cell line
[0082] (1) The HepG2 liver cancer cell line was cultured in DMEM-HG medium containing 10% FBS to a density close to 80%.
[0083] (2) Wash twice with PBS buffer to remove dead cells on the upper layer, add an appropriate amount of Trypsin to digest at 37°C for 3 minutes, then neutralize with DMEM-HG medium with 10% FBS, centrifuge and remove the supernatant.
[0084] (3) Resuspend in DMEM-HG medium with 10% FBS, count and passage, passage density: 1.0x10^5 / well.
[0085] (4) Preparation of the drug to be tested: Dilute the drug to an appropriate concentration, usually 10 mM. The test concentration is set to 1000x, 5000x, 10000x.
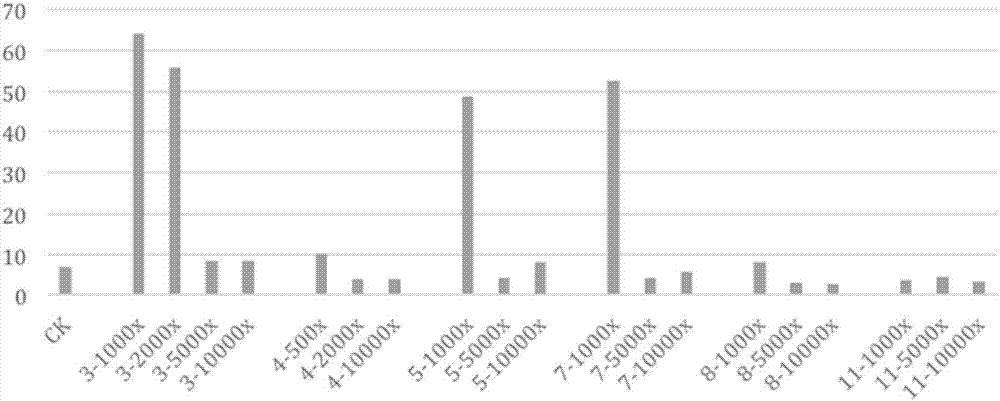
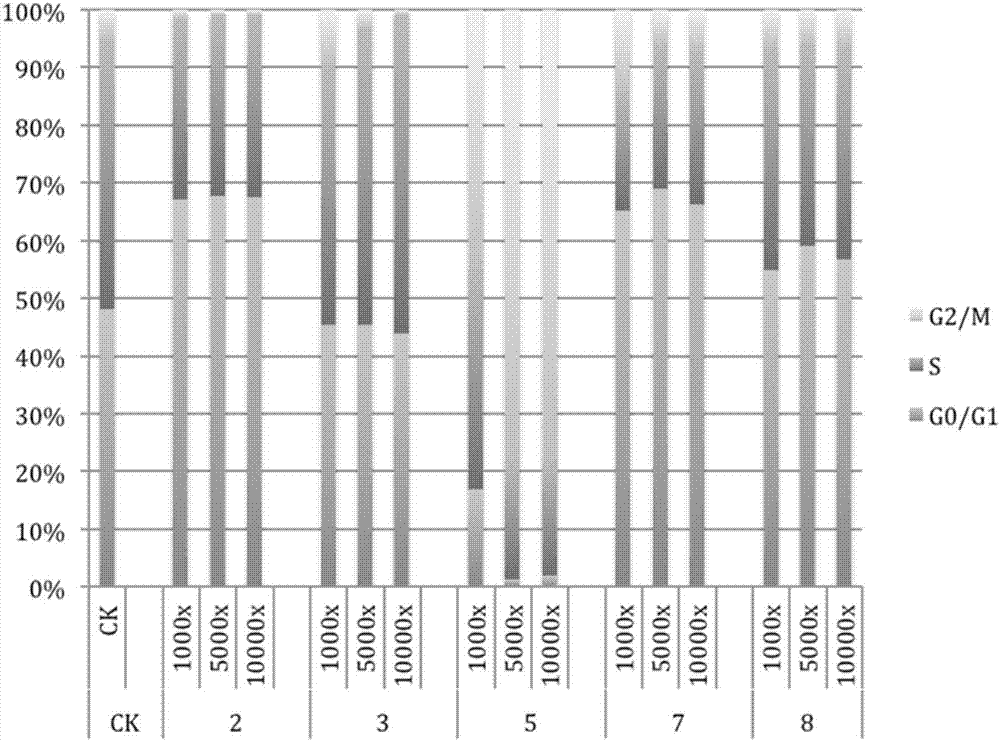
[0086] (5) After passage for 24 hours, add the drug to be tested when the confluence of the cells reaches about 60%. Three parallel wells were set up...
Embodiment 2
[0141] Example 2. Sequencing and result analysis of multi-label whole transcriptome sequencing library
[0142] (1) The library constructed in Example 1 was subjected to paired-end sequencing on the Illumina sequencing platform.
[0143] (2) Analyze the above sequencing results:
[0144] 1. Cluster analysis of sequencing results
[0145] Figure 5 It is the clustering analysis of the HepG2 gene expression of the drug to be tested for 72 hours. It can be seen that the targeted drug drug #2R428 (the target is Axl) and #5Crizotinib (the target is c-Met) have a lethal effect on liver cancer cell HepG2. similar expressive features. Unlike #7 Sunitinib (targeting RTK) and #3 Sorafenib (targeting ERK), a drug known to target liver cancer cells, it shows that their mechanisms of killing liver cancer cells HepG2 are different.
[0146] 2. Spantree analysis of sequencing results
[0147] Figure 6 It is the Spantree analysis of HepG2 gene expression of the drug to be tested for 72...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com