Method for site-specific integration of large DNA fragment in mammalian cell through CRISPR/Cas9
A technology of mammals and cells, applied in the field of genetic engineering, can solve problems such as unfavorable physical distance homologous recombination exchange, small exogenous DNA fragments, and low efficiency of double exchange homologous recombination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. CCR5 gene target site selection: According to the CCR5 gene sequence and the basic design principles of gRNA, and with the help of software analysis (http: / / crispr.mit.edu / ), design gRNA targeting the human CCR5 gene. The specific sequence is shown in Table 1 .
[0029] Table 1. Sequence information of gRNA targeting CCR5 gene
[0030] name Sequence (5'to 3') CRISPR / Cas9 Targeting Sites CATCCTGATAAACTGCAAA
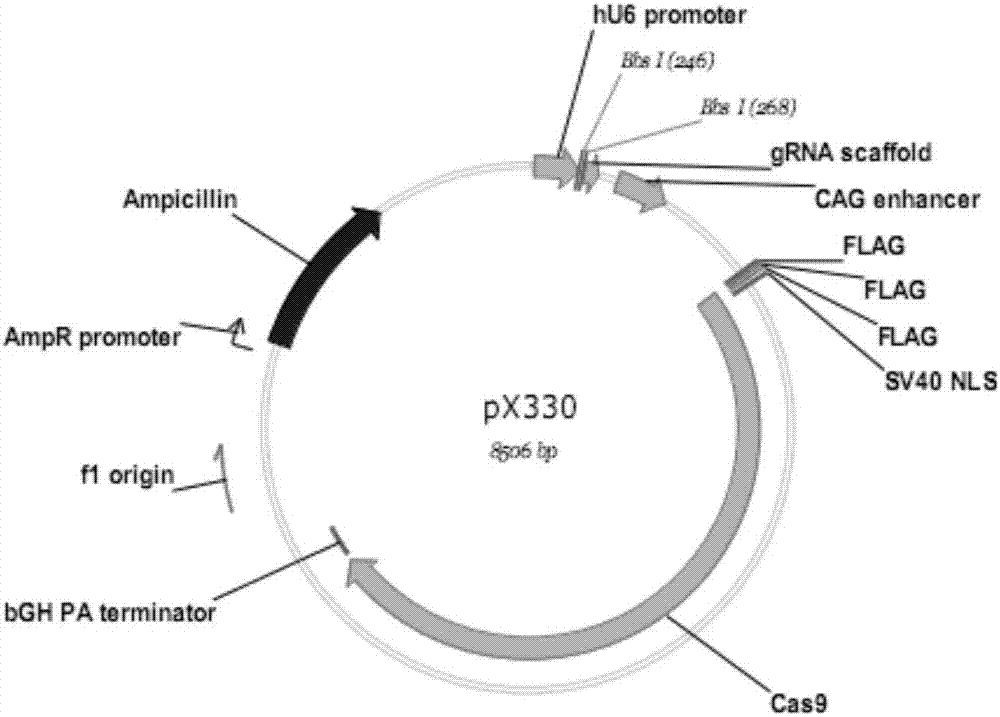
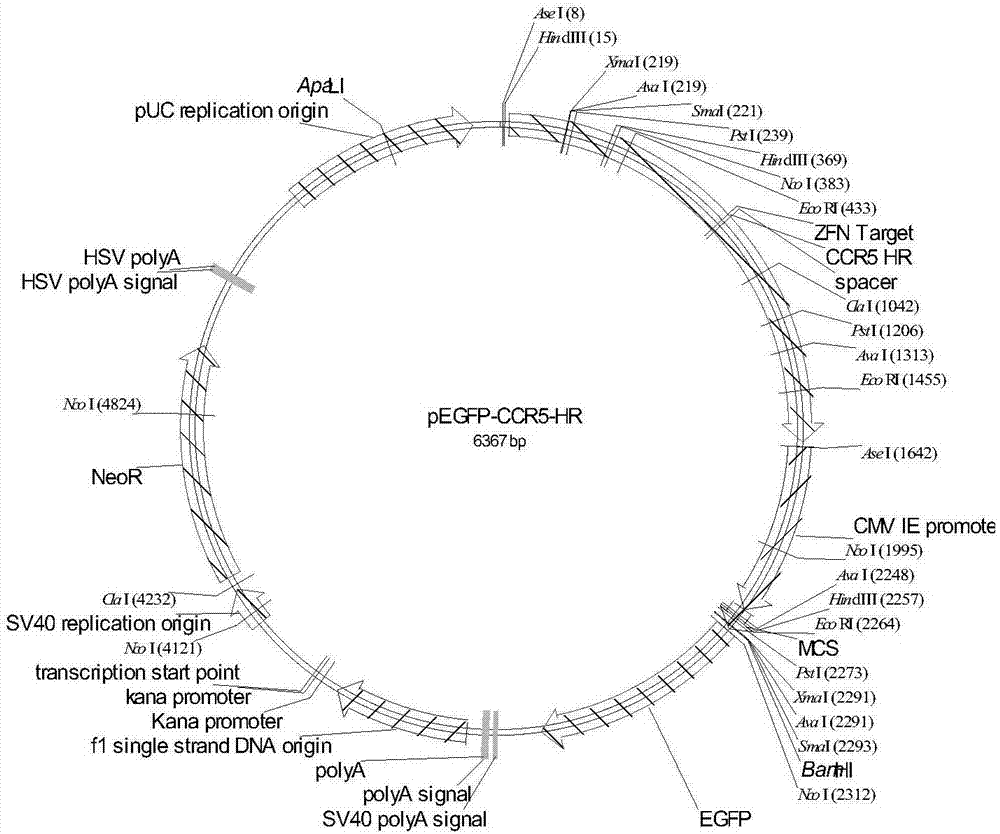
[0031] 2. HEK293T cell culture and transfection: DMEM high-glucose medium with 10% fetal bovine serum was used for HEK293T cell culture, and the cells were cultured in about 1×10 5 Cells / mL were inoculated in 24-well cell culture plates for use. When the confluence of the cells reaches about 90%, it can be used for transfection. For transfection, 150ng pX330-CCR5 plasmid, 100ngCCR5-EGFP-Donor plasmid (pEGFP-CCR5-HR), 1μL Lipofectamine 3000, 1μLP3000TM and 50μL Opti-MEM per well The medium formula was added for transfection, and a single-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com