Vaccines based on hepatitis b core antigens
A technology of hepatitis B and core antigen, which is applied in the field of vaccines based on hepatitis B core antigen, and can solve the problems that influenza vaccines have not yet been developed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0206] Materials and Methods:
[0207] VLP sequence:
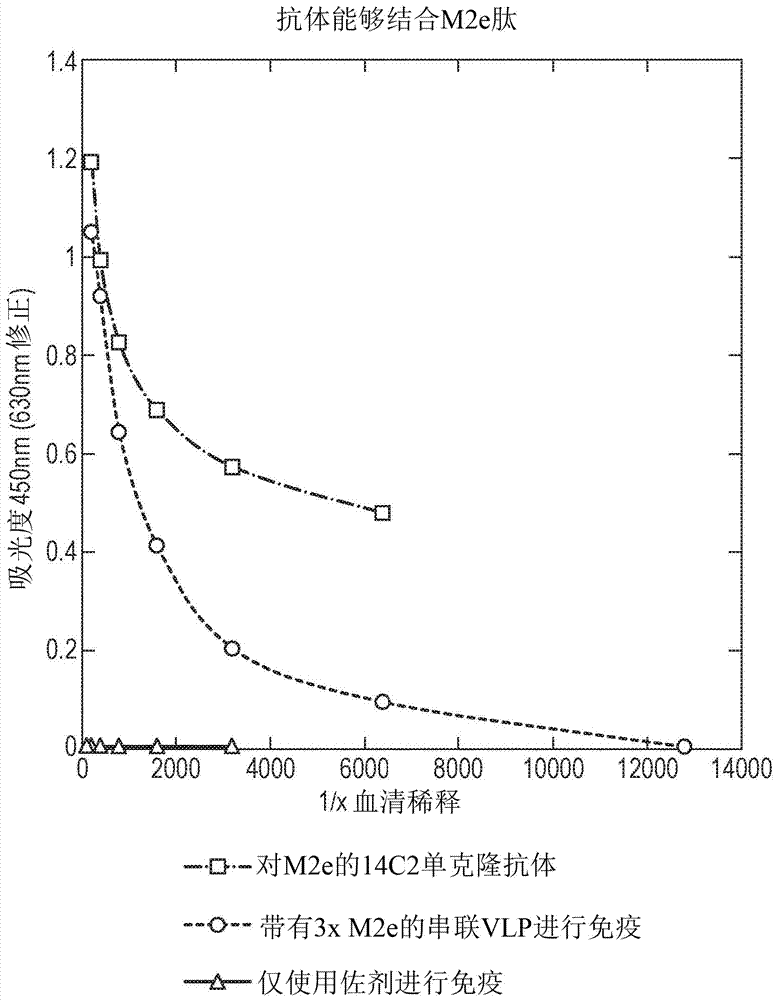
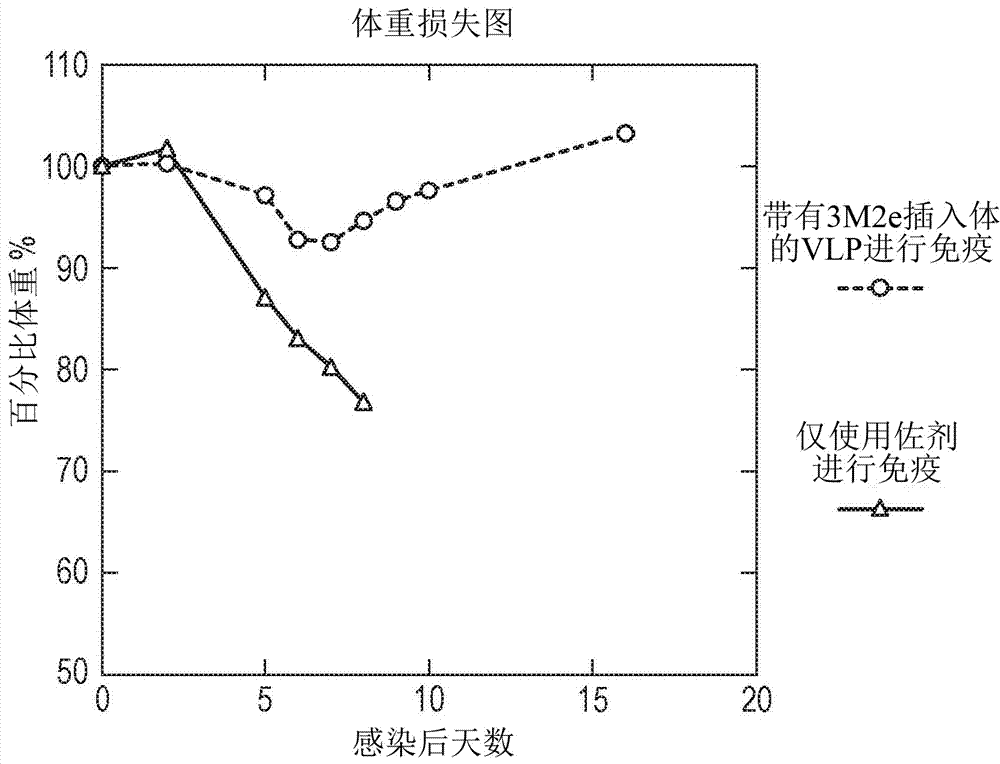
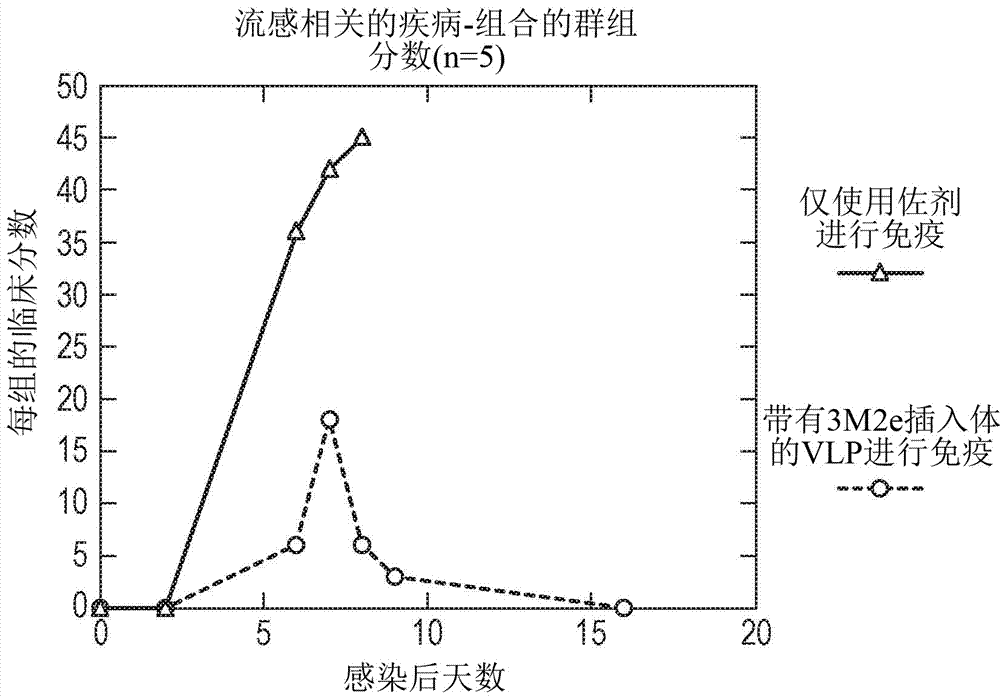
[0208] in BL21 E. coli A tandem core of inserts from influenza virus conserved proteins contained in MIRs was prepared in . The sequence corresponding to the insert belongs to the region of the extracellular domain of influenza virus matrix protein 2 (M2e). It spans 24 amino acids encoding the N-terminal external sequence of the known M2 protein ( Figure 5 ); MSLLTEVETPIRNEWG C R C NGSSD (SEQ ID NO: 6). The wild-type sequence was modified to replace cysteine residues (which affect VLP formation) at positions 17 and 19 (underlined) with serine residues. The final insert contained three variants of this sequence. The first version is the general M2e consensus sequence, except that the cysteine residues at positions 17 and 19 have been replaced by serine (SEQ ID NO: 9). The second (SEQ ID NO: 8) and third (SEQ ID NO: 10) are mutated versions of the general sequence, which correspond to the most common variants...
Embodiment 2
[0238] Materials and Methods:
[0239] VLP sequence:
[0240] in BL21 E. coli A tandem core of inserts from influenza virus conserved proteins contained in MIRs was prepared in . The sequence corresponding to said insert belongs to the stem region of the influenza virus hemagglutinin HA2 protein domain. It spans nucleic acid encoding known domains; loop B, helix C, helix CD, and helix D of the HA monomer ( Figure 10 ). The sequence spans amino acids (aa) 403-474 of the HA protein isolated from influenza A virus H1N1 / Lux / 09. There are other constructs comprising the HA stem insert sequence, these are described in Table 4 below.
[0241] Table 4. Table of alternative VLPs generated by changes in the insert sequence.
[0242]
[0243] The amino acid sequence of the tandem core with the HA stem influenza insert (encoded by the single letter aa) is shown below. Amino acids from the tandem core are bolded, while amino acids from influenza HA are underlined:
[0244]...
Embodiment 3
[0262] Materials and Methods:
[0263] VLP sequence:
[0264] in BL21 E. coli A tandem core containing insertions from conserved proteins of influenza virus within two major insertion regions (MIRs) was prepared in (Fig. 15). The sequence corresponding to the first insert belongs to the stem region of the influenza virus hemagglutinin HA2 protein domain. It spans nucleic acid encoding known domains; loop B, helix C, helix CD, and helix D of the HA2 monomer ( Figure 10 ). The sequence spans amino acids (aa) 403-474 of the HA protein isolated from influenza A virus H1N1 / Lux / 09. The sequence corresponding to the second insert belongs to the region of the extracellular domain of influenza virus matrix protein 2 (M2e). It spans 24 amino acids encoding the N-terminal external sequence of the known M2 protein; MSLLTEVETPIRNEWG C R C NGSSD (SEQ ID NO: 6) ( Figure 5 ). The wild-type sequence was modified to replace cysteine residues (which affect VLP formation) at posit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com