PCR (Polymerase Chain Reaction) detection method for mutation of human BRAF (v-raf Murine Sacoma Viral Oncogene Homolog) gene V600E, primer, probe and kit
A detection method and gene technology, applied in the field of detection of human BRAF gene V600E mutation status, can solve the problems of being difficult to apply to clinical detection, difficult to avoid cross-contamination, long detection period, etc., and achieve convenient and fast detection, fast and accurate detection. high sex effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1 designs and synthesizes specific primer and probe
[0047] According to the DNA sequence of the human BRAF gene published in the NCBI database, the upstream and downstream primers and probes for specifically detecting the human BRAF V600E site were designed. Among them, the 3' end of the upstream primer is a mutation site (c.1799T>A), and LNA is marked on this site.
[0048] According to the DNA sequence of the human GAPDH gene published in the NCBI database, the upstream and downstream primers and probes for specific detection of the human GAPDH gene were designed.
[0049] The nucleotide sequences (5'-3') of the designed 4 primers and 2 probes are as follows:
[0050] BRAF upstream primer: GTGATTTTGGTCTAGCTACAG+A (SEQ ID NO: 1)
[0051] BRAF downstream primer: ATCCAGACAACTGTTCAAACTGATG (SEQ ID NO: 2)
[0052] BRAF probe: FAM-CTCGATGGAGTGGGTC-MGB (SEQ ID NO: 3)
[0053] GAPDH upstream primer: CTGCCAGCCTAGCGTTGAC (SEQ ID NO: 4)
[0054] GAPDH downstrea...
Embodiment 2
[0057] Specificity, sensitivity and linear regression experiment (based on fluorescence quantitative PCR platform) of embodiment 2 primers and probe
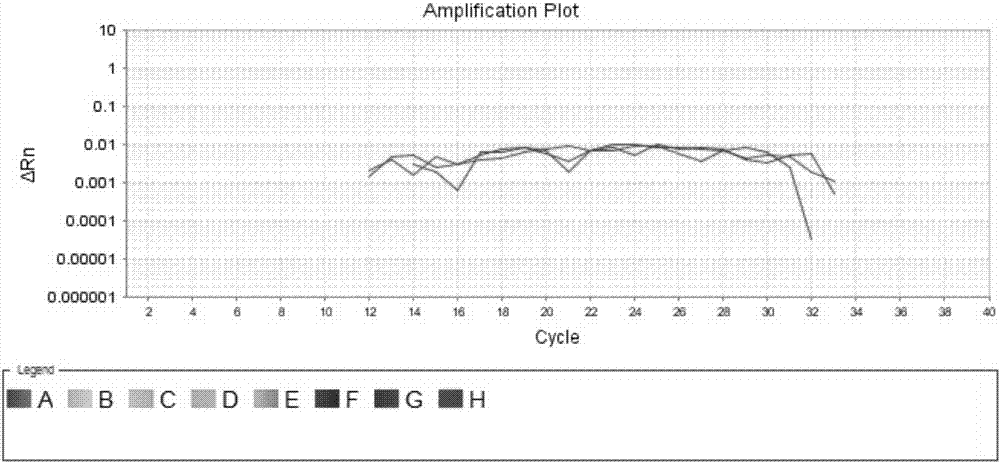
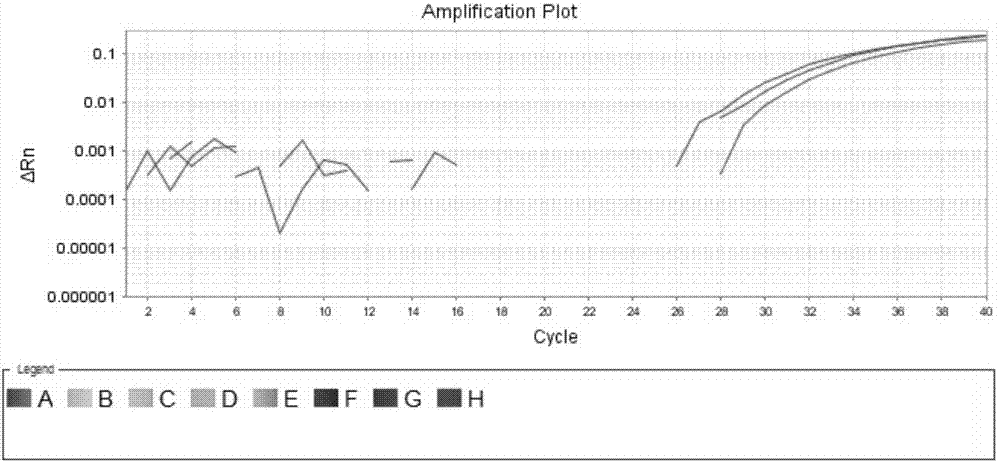
[0058] Genomic DNA containing only BRAF gene V600E wild-type and genomic DNA containing only BRAF gene V600E mutant were used as templates, and the specificity and sensitivity of the primers and probes for the above-mentioned BRAF gene V600E locus were detected by fluorescent quantitative PCR platform. The detection system is shown in Table 1:
[0059] Table 1 Specificity and Sensitivity Detection System of Primers and Probes (Fluorescence Quantitative PCR Platform)
[0060] Element
Volume (μL)
MgCl 2
0.4
KCl
2
(NH4) 2 SO 4
2
Tris-Hcl
2
ROX II
0.2
Taq enzyme
0.2
BRAF-F
0.4
BRAF-R
0.4
GAPDH-F
0.4
GAPDH-R
0.4
BRAF probe
0.3
GAPDH probe
0.3
template+water
11
total capacity
2...
Embodiment 3
[0078] Specificity, sensitivity and linear regression experiment (based on digital PCR platform) of embodiment 3 primers and probe
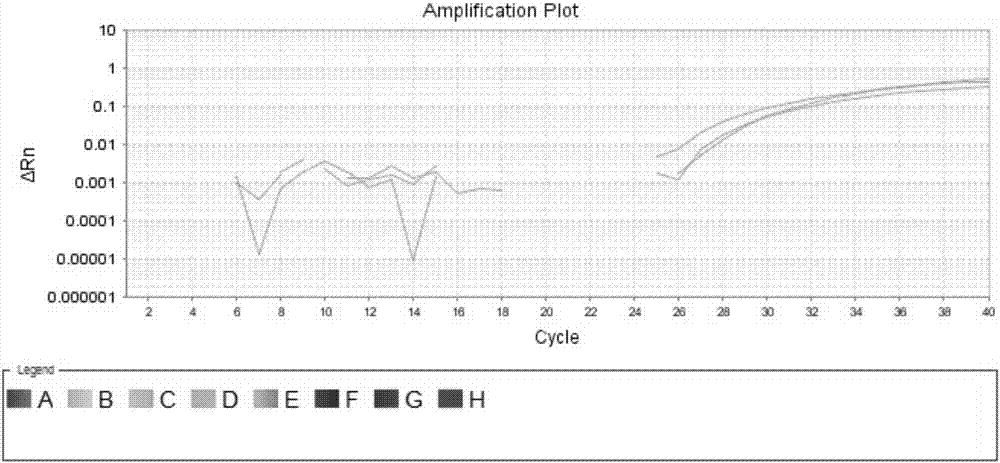
[0079] Genomic DNA containing only BRAF gene V600E wild-type and genomic DNA containing only BRAF gene V600E mutant were used as templates, respectively, using a digital PCR platform to detect the specificity and sensitivity of the primers and probes for the BRAF gene V600E site. The detection system is shown in Table 5:
[0080] Specificity and sensitivity detection system of table 5 primers and probes (digital PCR platform)
[0081] Element
Volume (μL)
2*ddPCR Supermix for Probes
10
BRAF-F
1.8
BRAF-R
1.8
GAPDH-F
1.8
GAPDH-R
1.8
BRAF probe
0.5
GAPDH probe
0.5
template+water
1.8
wxya 2 o
20
[0082] The detection method of specificity and sensitivity is as follows: prepare the digital PCR detection system according to Table 5, then trans...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com