Methods for tagging dna-encoded libraries
A coding and library technology, applied in the direction of DNA preparation, recombinant DNA technology, chemical library, etc., can solve problems such as inability to shift
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
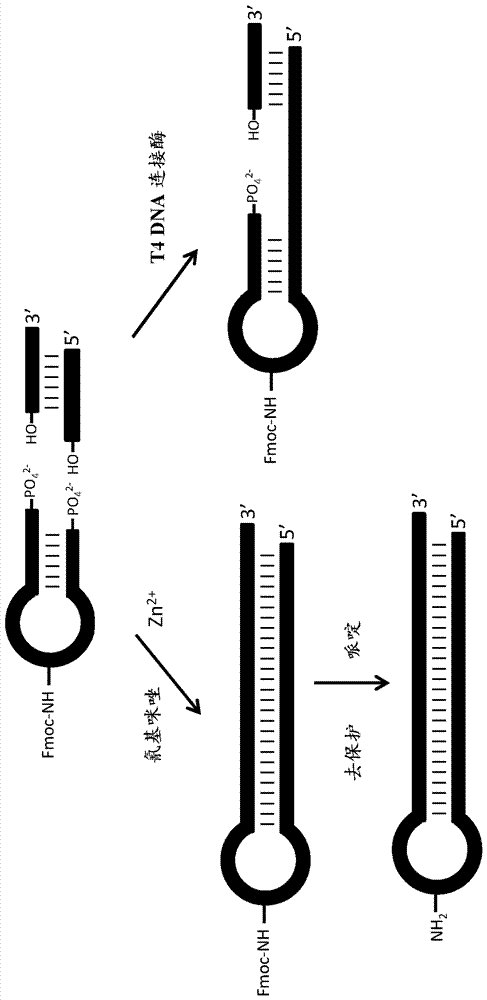
[0206] Example 1. Preparation of components for chemical ligation (double-stranded head fragment and double-stranded label)
[0207] The head fragment HP006 chemically phosphorylated at the 5' end, SEQ ID NO: 1 -(p)CCTGTGTTZTTCACGGCCT, where Z represents C6-amino dT modification, was obtained from Biosearch Inc. HP006 was subsequently modified by DMT-MM acylation with Fmoc-NH-PEG4-CH2CH2COOH (Chem Pep Inc) using the following method.
[0208] 50 equivalents of Fmoc-NH-PEG4-CH2CH2COOH (Chem Pep Inc) were dissolved in DMA (dimethylacetamide, Acros), and 1 equivalent of HP006 dissolved in 0.5M borate buffer at pH 9.5 was added and 50 equivalents of DMT-MM (4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholine hydrochloride, Acros) freshly dissolved in water . The reaction was allowed to proceed for 2-4 hours, followed by a second addition of 50 equiv of Fmoc-NH-PEG4-CH2CH2COOH and 50 equiv of DMT-MM, and then the reaction was allowed to proceed overnight. The reaction was mo...
Embodiment 2
[0214] Embodiment 2: chemical connection of double-stranded head fragment and double-stranded label
[0215] Fmoc-amino-PEG4-HP013 and double-stranded TagZA oligonucleotides were dissolved in 80 mM MES buffer (containing 800 mM NaCl and 8 mM ZnCl 2 ) to a final concentration of 0.33 mM. 1-cyanoimidazole was freshly dissolved in DMF at a concentration of 1 M, and 1-2 additions were made to the reaction over 12 hours to a final concentration of 1-cyanoimidazole of 150 mM. Reactions were then incubated overnight at 4°C.
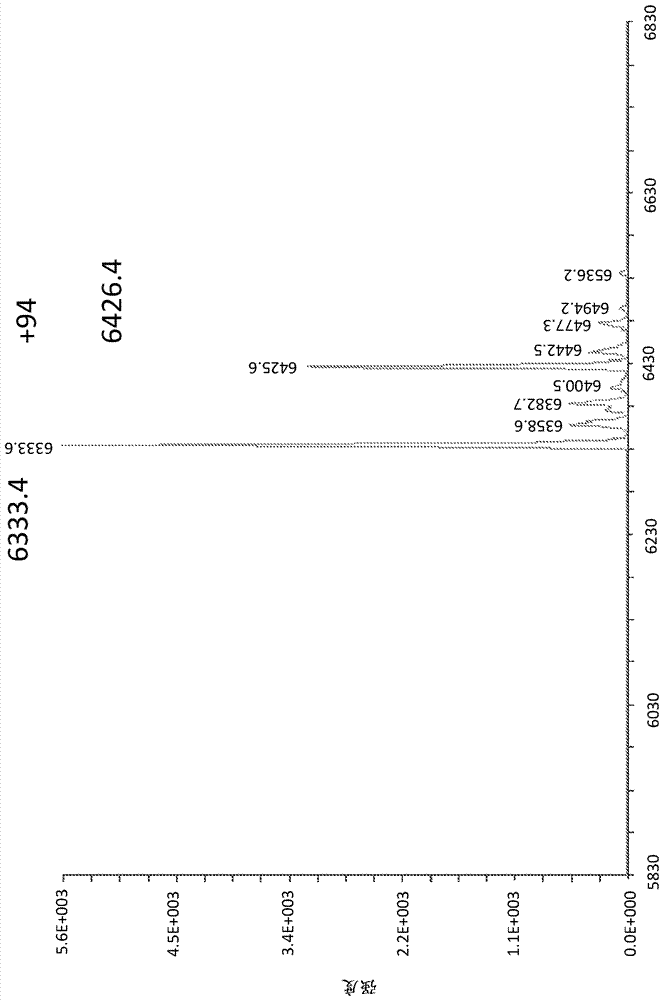
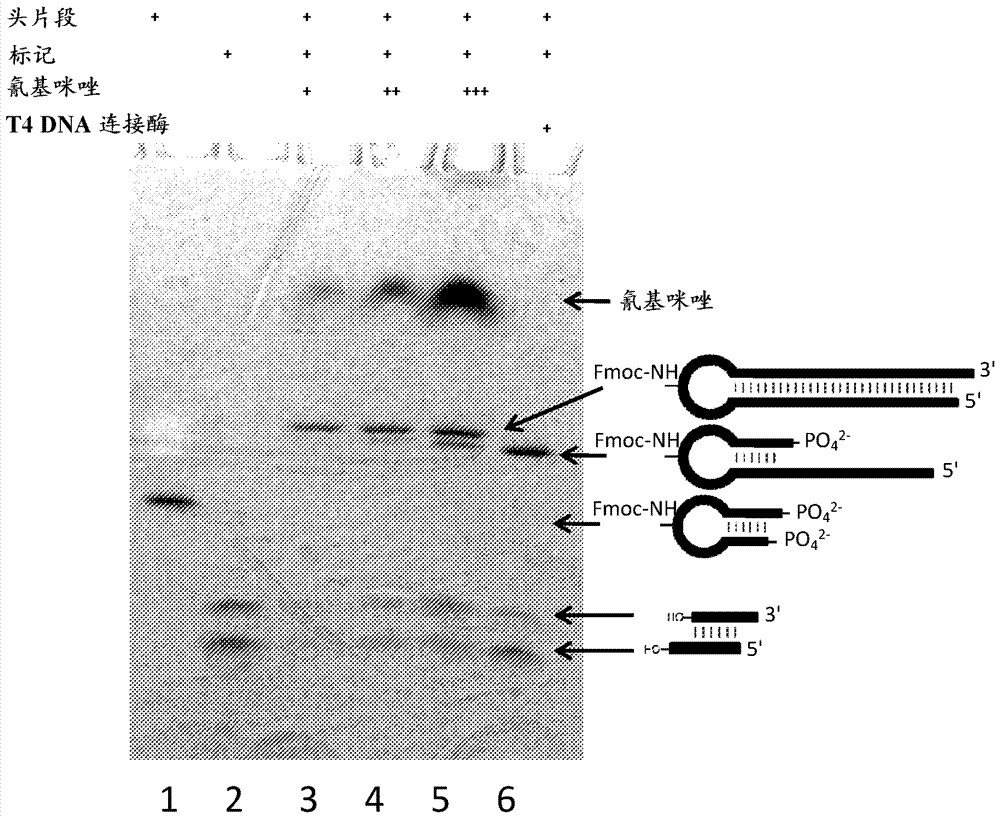
[0216] Completed reactions were analyzed by denaturing gel electrophoresis and LCMS. Samples were then separated in a 15% denaturing analytical TBE-8M urea gel and visualized on TLC plates by UV shadowing using fluorescent dyes (254 nm). LCMS confirmed the formation of a double-stranded ligated product with MW 25,417.3 (calculated molecular weight 25,415.3 ) and ~70% conversion. Additional products of MW 20,254.7 and 18,935.4 were observed, corresponding to ...
Embodiment 3
[0225] Example 3. Fmoc deprotection of chemical ligation reaction products.
[0226]The product of the 1-cyanoimidazole ligation reaction was ethanol precipitated, dissolved in water and deprotected by incubation in 10% piperidine for 2 hours at room temperature. After this deprotection step, the material was purified on a 15% TBE-8M urea gel. LC-MS on the purified sample confirmed the presence of deprotected amino-PEG4-HP013-TagZA (MW 25,192.4, calculated molecular weight 25,193.2) and two semi-linked deprotected products (MW 18,738.6 and 20,029.3).
[0227] Integration of the LC traces gave a relative yield of 64% for the full-length product and about 18% for each of the hemiligated products. The ligation efficiency of each strand was predicted to be 83%.
[0228] Deprotection of the amino group by piperidine as Figure 4A shown. Gel purification of ligation reaction products: 15% TBE-urea gel, UV contrast such as Figure 4B shown. LCMS analysis of purified material as...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com