Method of analyzing and quantifying thallus quantity of probiotics and pathogenic bacteria by means of molecular biological technique
A technology of molecular biology and technical analysis, which is applied in the field of analyzing and quantifying the number of probiotics and pathogenic bacteria with molecular biology techniques, and can solve the problems of probiotic pathogenic bacteria detection, culture counting, lack of reasonableness, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Embodiment 1: the quantitative detection method of Lactobacillus paracasei (L.paracasei) in commercially available probiotic capsule product
[0066] Step 1. Pre-processing:
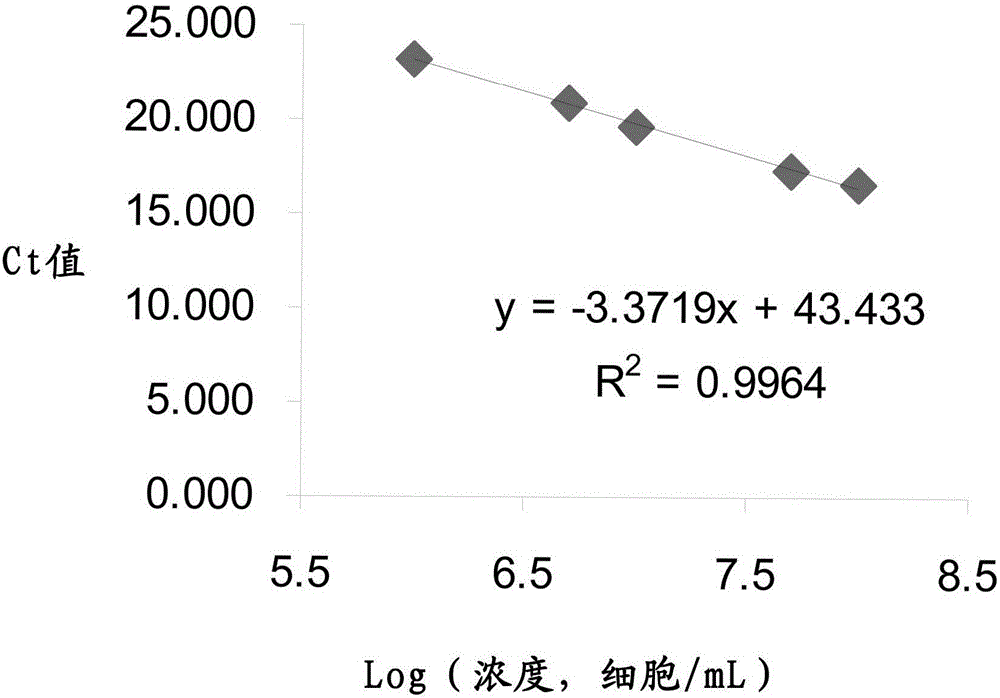
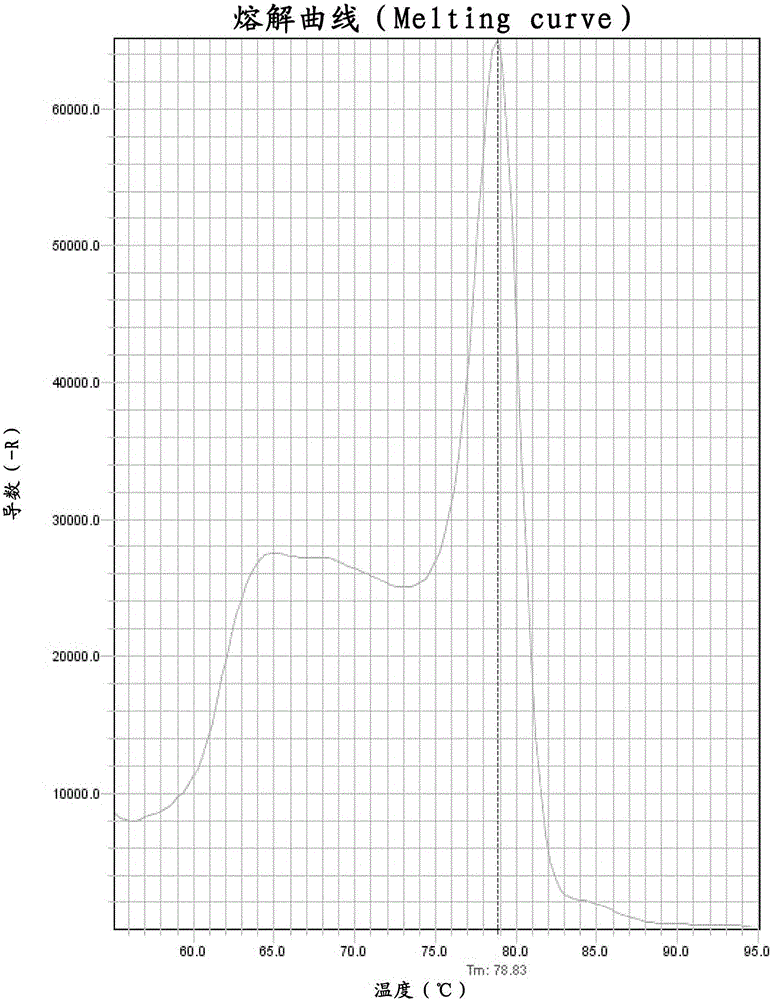
[0067] Take the commercially available capsule product containing Lactobacillus paracasei (strain registration number CGMCC No.3247), take out the tested bacterial powder in the capsule, weigh 1±0.005g and place it in a sterile centrifuge tube, and then add appropriate amount of sterile Water Use a shaker to fully dissolve the tested bacteria powder in sterile water. Centrifuge at 10,000-11,000rpm and 4°C for 30 minutes. After centrifugation, remove the supernatant, add sterile water to lyse the bacteria in a small amount and multiple times, pour into a suitable quantitative bottle and perform quantitative determination. After dilution to an appropriate multiple, the Ct value of the test product can fall in the description of the attached drawing figure 1 within the range of the standard curve. ...
Embodiment 2
[0077] Embodiment 2: Quantitative detection method of Lactobacillus fermentum in probiotic powdery product
[0078] Step 1. Pre-processing:
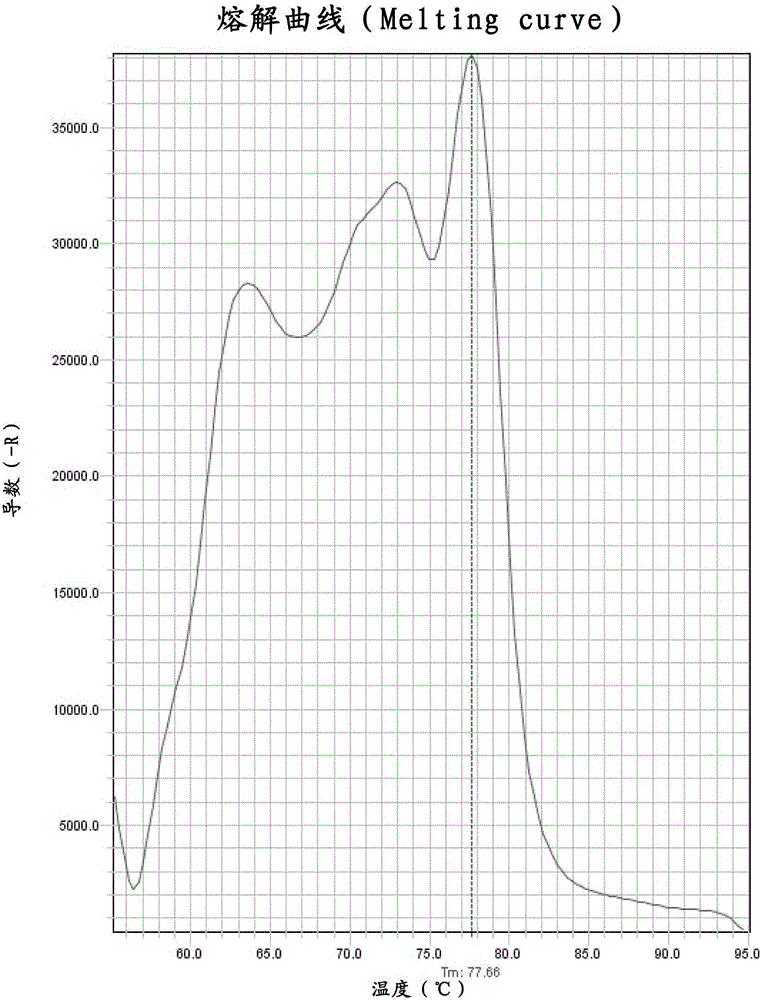
[0079] Weigh 1±0.005 g of the probiotic powder product containing Lactobacillus fermentum (L.fermentum) (strain registration number CGMCCNo.3248) into a sterile centrifuge tube, then add an appropriate amount of sterile water and shake it with a shaker. The tested bacteria powder was fully dissolved in sterile water. Centrifuge at 10,000-11,000rpm and 4°C for 30 minutes. After centrifugation, remove the supernatant, add sterile water to lyse the bacteria in a small amount and multiple times, pour into a suitable quantitative bottle and perform quantitative determination. After dilution to an appropriate multiple, the Ct value of the test product can fall in the description of the attached drawing Figure 4 within the range of the standard curve.
[0080] Step 2. Real-time Quantitative Polymerase Chain Reaction:
[0081] Using the spe...
Embodiment 3
[0090] Example 3: Quantitative detection method of individual strains in mixed probiotic products
[0091] Mixed probiotic products individually contain the same proportion of Lactobacillus paracasei and Lactobacillus fermentum, the number of which is 5.6×10 11 cells / g and 1.8×10 11 cells / g, and if the traditional spectrophotometer is used to detect the number of bacteria at a wavelength of 600nm for mixed probiotic products, the total number of bacteria can only be obtained from the concentration of bacterial cells, and the number of individual bacteria cannot be known. Amplified by a strain-specific primer set, it can accurately detect the quantification of the number of individual strains (Lactobacillus paracasei and Lactobacillus fermentum) in mixed probiotic products.
[0092] Step 1. Pre-processing:
[0093] Weigh 1±0.005 g and place it in a sterile centrifuge tube, then add an appropriate amount of sterile water and fully dissolve the tested bacterial powder in steril...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com