Molecular marker linked to introgression line's stripe rust resistance site from Agropyron P genome in adult-plant stage and application thereof
A technology for molecular markers and stripe rust, which is applied in special data processing applications, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of general limitation, marker target gene far away, and few applications, so as to speed up the breeding process , close genetic distance and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Obtaining the SNAP molecular marker 2B_SNAP_102 linked to the resistance gene of adult plant stage of stripe rust from the introgression line of wheatgrass P genome
[0029] 1. Test materials
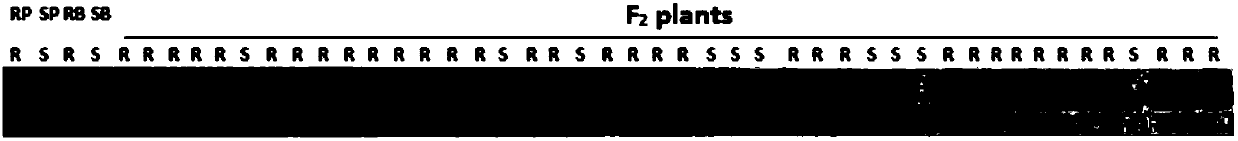
[0030] Plant materials: the adult plant resistant wheat-Agrocybe introgression line Pubing 3558 was used as the female parent, and the susceptible variety Avocet S was used as the male parent to construct F 2 Genetic Mapping Populations. Planting will F 2 152 individual plants from the population were used for resistance identification. Some leaves of each individual plant were taken at the seedling stage for SNAP analysis. When ripe, harvest F 2:3 The seeds of the family were dried and preserved for resistance identification.
[0031] Physiological race of stripe rust: for F 2 group and F 2:3 The physiological race of P. stripe rust identified by family resistance is the purified physiological race 33 in Tiaozhong.
[0032] 2. Identification of resistance to s...
Embodiment 2
[0047] Example 2: Molecular-assisted selection of wheat stripe rust resistance gene at adult plant stage
[0048] 1. Test materials
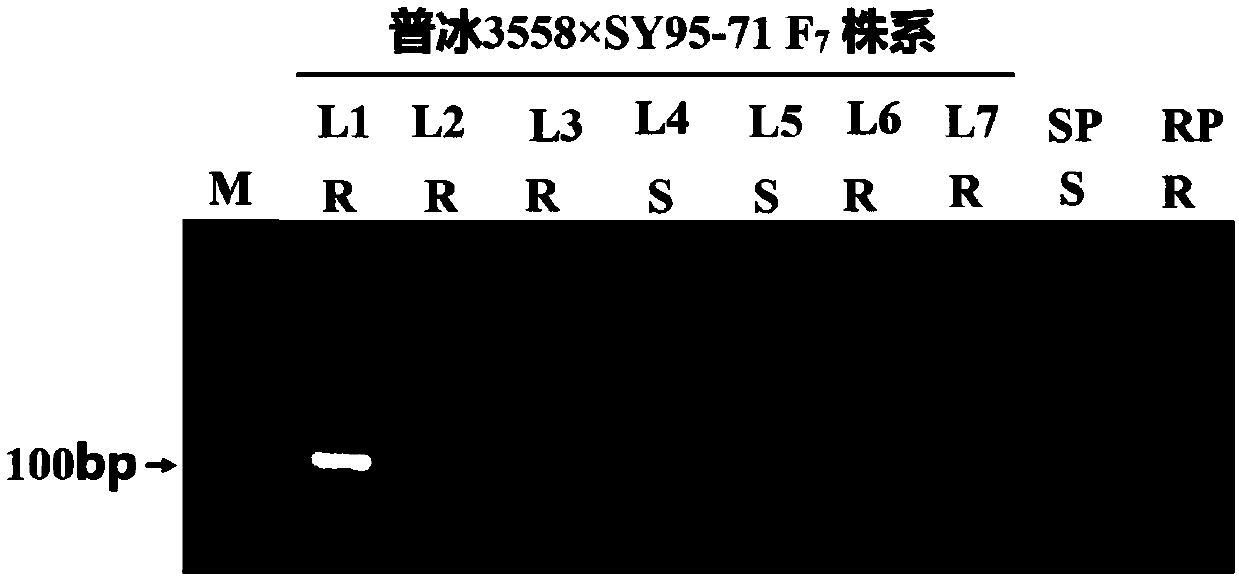
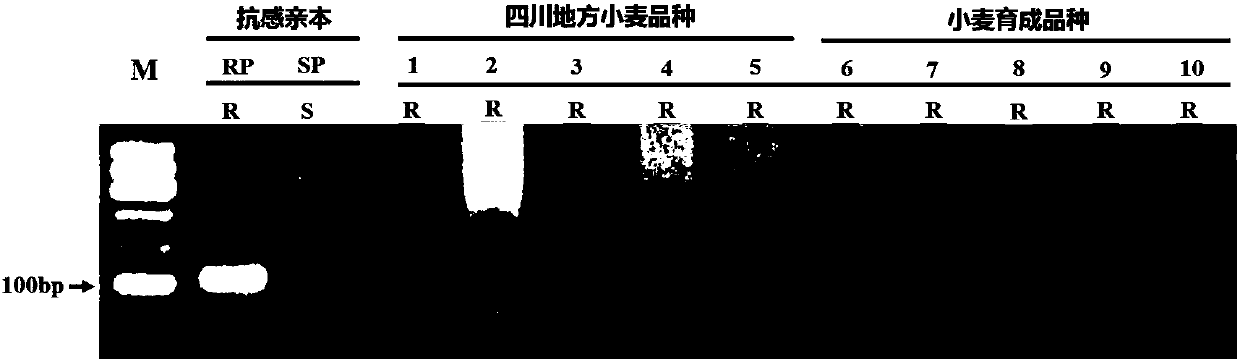
[0049] Plant materials: 7 wheat lines (L1-L7) of the advanced generation (F7) of the hybrid combination of the disease-resistant parent Pubing 3558 and the susceptible line SY95-71, 5 disease-resistant Sichuan local wheat varieties (Lezhihong Tiaomai, Deyangtian Lvchang wheat, Gusong Dalongxu, Fengdu Pushanmai, Zhongjiang Aizi Huoshaomai) and 5 wheat cultivars unrelated to the disease-resistant parent Pubing 3558 (Chuanmai 33, Chuanmai 61, Mianmai Mai 41, Mianmai 1403, Zhongkemai 47) The above materials were planted in the Chongzhou Experimental Base of Sichuan Agricultural University in the first season of 2016 for the identification of stripe rust resistance in adult plants. Some leaves of each individual plant were taken at the seedling stage for SNAP analysis.
[0050] Mixed physiological races of stripe rust: The physiological races of st...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com