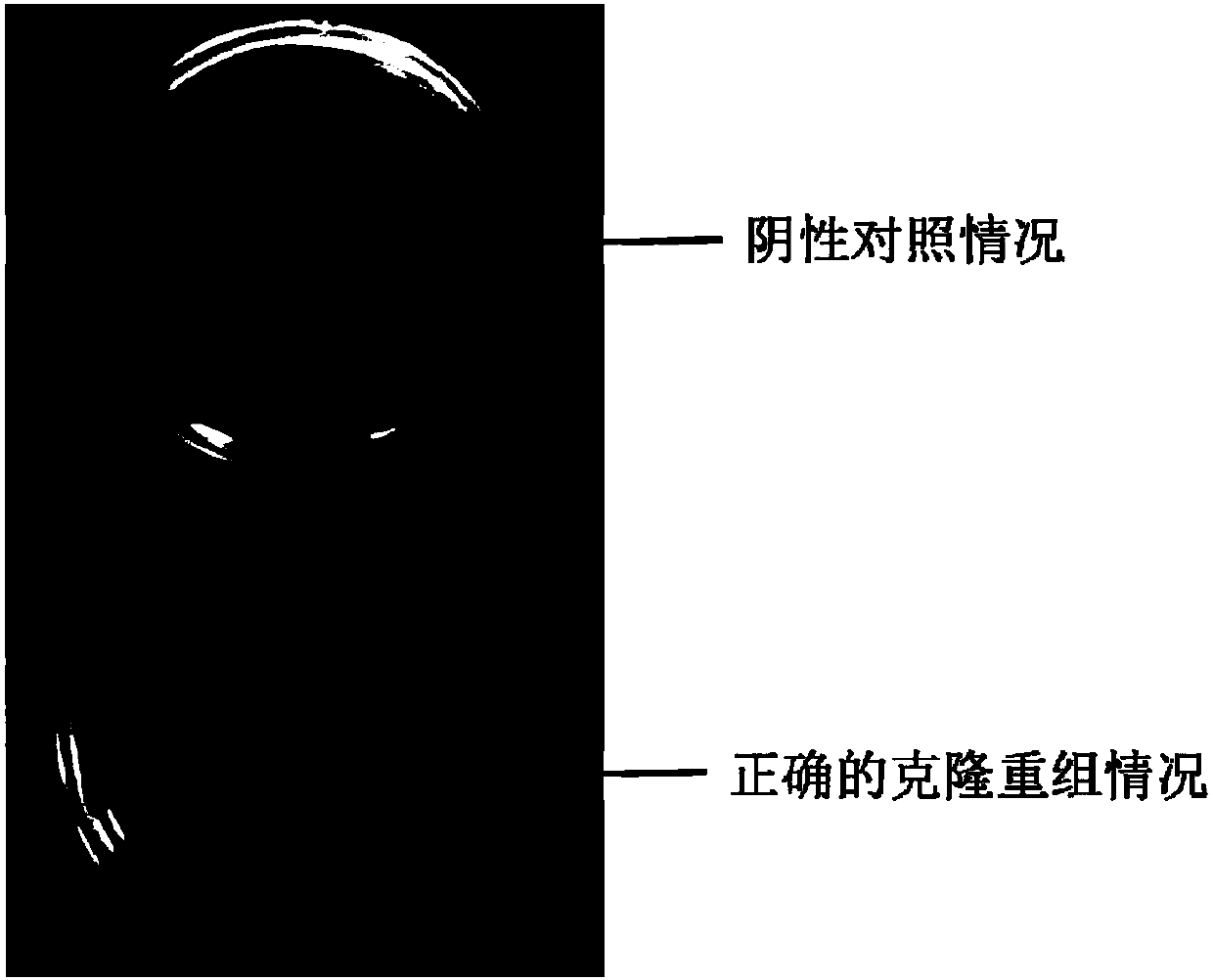
Linearized CRISPR-CAS9 lentiviral vector-based gene knockout kit and application thereof
A technology of lentiviral vectors and kits, applied in applications, genetic engineering, plant genetic improvement, etc., can solve problems such as lack of kits, achieve the effects of eliminating inconsistencies, avoiding differences in connection efficiency, and reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 (CRISPR-CAS9 lentiviral vector gene knockout with puromycin resistance constructed by common cloning and recombination method, taking SNX10 gene as an example)
[0054] 1. Mouse SNX10 gene sgRNA design:
[0055] (1) Design the positive and antisense strand primer sequences of the SNX10 gene:
[0056] sgRNA1F: AAACGCAACGCATTACTTTGGAGTC SEQ ID No.1
[0057] sgRNA1R: CACCGCACGTGGATCAGCGTCGCCA SEQ ID No. 2
[0058] sgRNA2F: CACCGCACGTGGATCAGCGTCGCCA SEQ ID No.3
[0059] sgRNA2R: AAACTGGCGACGCTGATCCACGTGC SEQ ID No.4
[0060] (2) The linearized CRISPR-CAS9 lentiviral vector with puromycin resistance described in the kit;
[0061] (3) T4 ligase and T4 ligase buffer;
[0062] (4) High transformation activity Stbl3 competent;
[0063] 2. Recombination of CRISPR-CAS9 lentiviral cloning vector with puromycin resistance
[0064] (1) Anneal the sense strand and the antisense strand to form double strands: take equal amounts of the sense strand and the antisense str...
Embodiment 2
[0080] Example 2 (CRISPR-CAS9 lentiviral vector gene knockout with puromycin resistance constructed by common cloning and recombination method, taking HSF1 gene as an example)
[0081] 1. Human HSF1 gene sgRNA design:
[0082] (1) Design the positive and antisense strand primer sequences of the HSF1 gene:
[0083] sgRNA1F: TCGTGAGCGACCCGGACACCGTTTT SEQ ID No.5
[0084] sgRNA1R: GGTGTCCGGGTCGCTCACGACGGTG SEQ ID No. 6
[0085] sgRNA2F: CAGCTTCCACGTGTTCGACCGTTTT SEQ ID No. 7
[0086] sgRNA2R: GGTCGAACACGTGGAAGCTGCGGTG SEQ ID No.8
[0087] sgRNA3F: GGAGTCAATGAGGGCGGTCGGTTTT SEQ ID No. 9
[0088] sgRNA3R: CGACCGCCCTCATTGACTCCCGGTG SEQ ID No.10
[0089] (2) The linearized CRISPR-CAS9 lentiviral vector with puromycin resistance described in the kit;
[0090] (3) T4 ligase and T4 ligase buffer;
[0091] (4) High transformation activity Stbl3 competent;
[0092] 2. Recombination of CRISPR-CAS9 lentiviral cloning vector with puromycin resistance
[0093] (1) Anneal the sense st...
Embodiment 3
[0109] Example 3 (CRISPR-CAS9 lentiviral vector gene knockout with puromycin resistance constructed by common cloning and recombination method, taking PKA gene as an example)
[0110] 1. Rat PKA gene sgRNA design:
[0111] (1) Design and obtain the positive and antisense strand primer sequences of the PKA gene:
[0112] sgRNA1F: AACGCCGCCGCCGCCAAGAAGTTTT SEQ ID No.11
[0113] sgRNA1R: TTCTTGGCGGCGGCGGCGTTCGGTG SEQ ID No.12
[0114] sgRNA2F: CCTCCCAATCCGCCGTAAGTGTTTT SEQ ID No. 13
[0115] sgRNA2R: ACTTACGGCGGATTGGGAGGCGGTG SEQ ID No.14
[0116] sgRNA3F: CGATCTGCGCCGCGTAGAAAGTTTT SEQ ID No. 15
[0117] sgRNA3R: TTTCTACGCGGCGCAGATCGCGGTG SEQ ID No. 16
[0118] (2) The linearized CRISPR-CAS9 lentiviral vector with puromycin resistance described in the kit;
[0119] (3) T4 ligase and T4 ligase buffer;
[0120] (4) High transformation activity Stbl3 competent;
[0121] 2. Recombination of CRISPR-CAS9 lentiviral cloning vector with puromycin resistance
[0122] (1) Anneal the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com