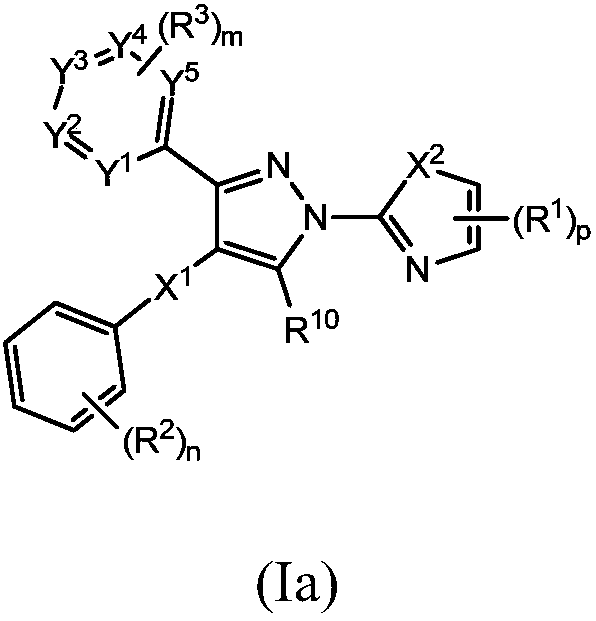
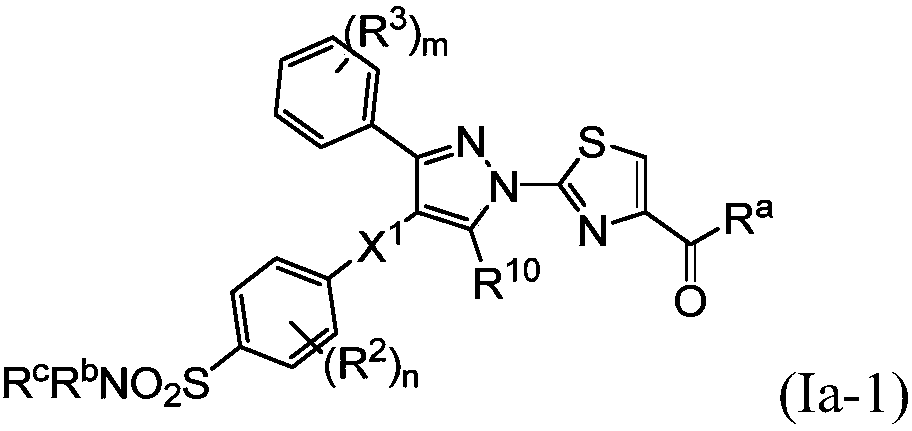
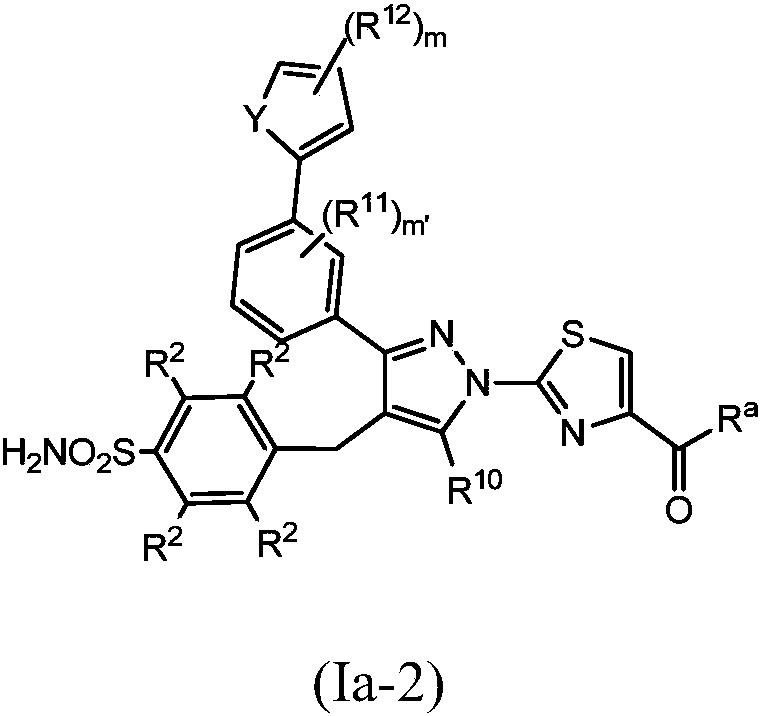
Small molecule inhibitors of lactate dehydrogenase and methods of use thereof
A heteroatom and hydrocarbon-based technology, applied in the field of small molecule inhibitors of lactate dehydrogenase and its application, can solve the problems of poor bioavailability of inhibitors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0299] This example describes the human LDHA primary biochemical assay used to characterize compounds of formula (I) in an embodiment of the invention.
[0300] Test compounds were placed in Greiner Bio-One (Monroe, NC) 1536-well black solid bottom assay plates. 200 millimolar (mM) Tris HCl, pH 7.4, 100 micromolar (μM) EDTA and 0.01% TWEEN-20 TM , final concentration, used as assay buffer. LDHA reagent was 2 nanomolar (nM) human LDHA (Meridian Life Science, Inc., Memphis, TN), final concentration, in assay buffer. Substrate reagents were 0.06 mM NADH and 0.2 mM sodium pyruvate, final concentrations, in assay buffer. The resazurin / diaphorase coupling reagent was 0.037 mM resazurin and 0.133 milligrams per milliliter (mg / mL) diaphorase in assay buffer, final concentration. The sequence of steps, the amount and type of reagents required for each step and the timing are listed in Table 1. Inhibition of LDHA activity was measured by fluorescence emission.
[0301] Table 1
[...
Embodiment 2
[0304] This example describes the human LDHB enumeration screening biochemical assay used to characterize compounds of formula (I) in an embodiment of the invention.
[0305] Test compounds were placed in Greiner Bio-One (Monroe, NC) 1536-well black solid bottom assay plates. 200 mM Tris HCl, pH 7.4, 100 μM EDTA and 0.01% TWEEN-20 TM , final concentration, used as assay buffer. LDHB reagent was 2 nM human LDHB (Meridian Life Science, Inc., Memphis, Tenn.), final concentration, in assay buffer. Substrate reagents were 0.13 mM NADH and 0.16 mM sodium pyruvate, final concentrations, in assay buffer. The resazurin / diaphorase coupling reagent is 0.037 mM resazurin and 0.133 mg / mL diaphorase, final concentration, in assay buffer. The sequence of steps, the amount and type of reagents required for each step and the timing are listed in Table 2. Inhibition of LDHB activity was measured by fluorescence emission.
[0306] Table 2
[0307]
Embodiment 3
[0309] This example describes the human PHGDH enumeration screening biochemical assay used to characterize compounds of formula (I) in an embodiment of the invention.
[0310] Test compounds were placed in Greiner Bio-One (Monroe, NC) 1536-well black solid bottom assay plates. 50mM TEA, pH 8.0, 10mM MgCl 2 , 0.05% BSA and 0.01% TWEEN-20 TM , the final concentration was used as the assay buffer. Substrate reagents were 10 μM EDTA, 0.625 mM glutamate, 500 nM human PSAT1, 500 nM human PSPH, 0.05 mM 3-phosphoglycerate kinase, 0.1 mM resazurin, and 0.1 mg / mL diaphorase, final concentrations, in assay buffer. PHGDH reagent is 0.15mM NAD + and 10 nM human PHGDH, final concentration, in assay buffer. The sequence of steps, the amount and type of reagents required for each step and the timing are listed in Table 3. Inhibition of PHGDH activity was measured by fluorescence emission.
[0311] table 3
[0312]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com