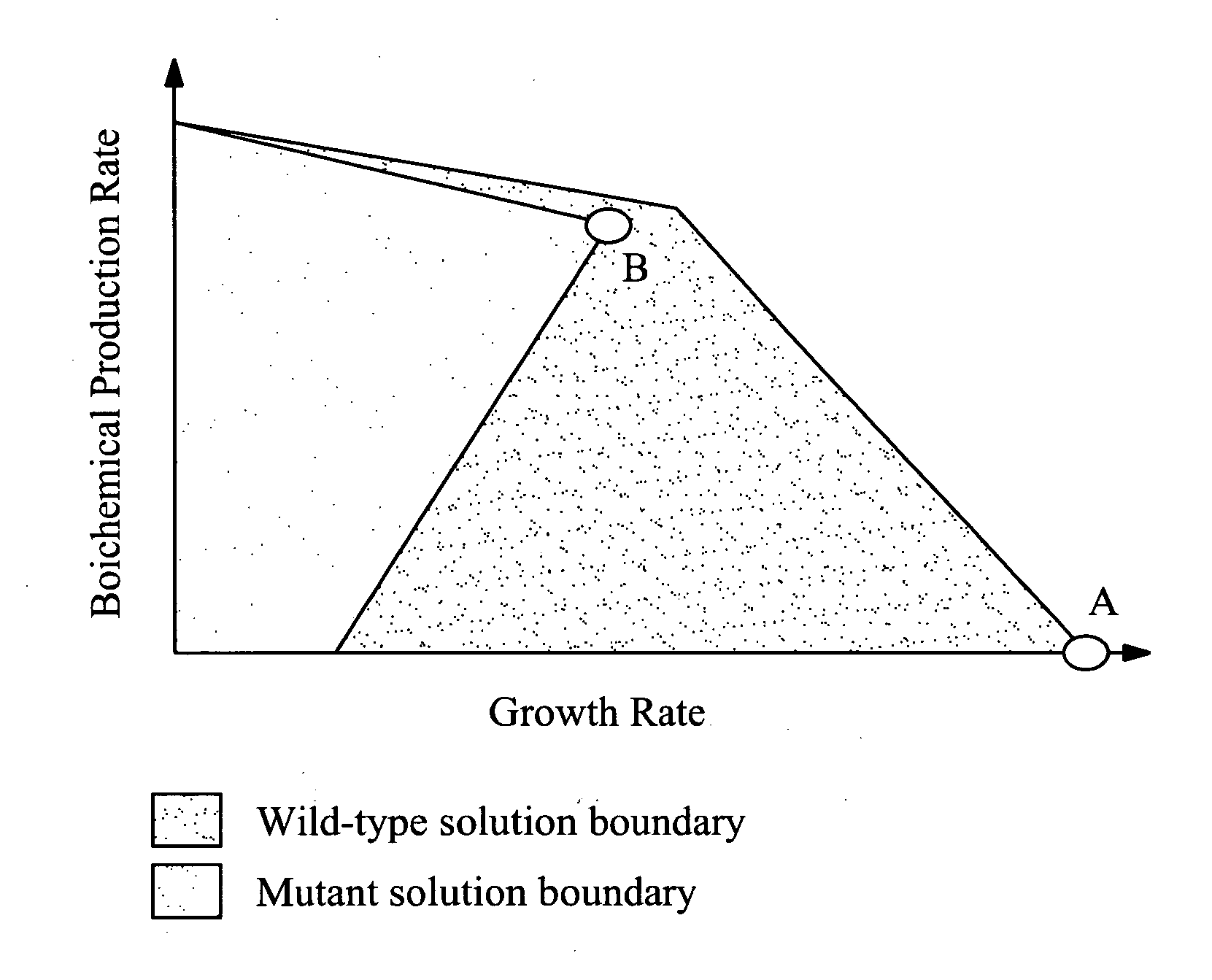
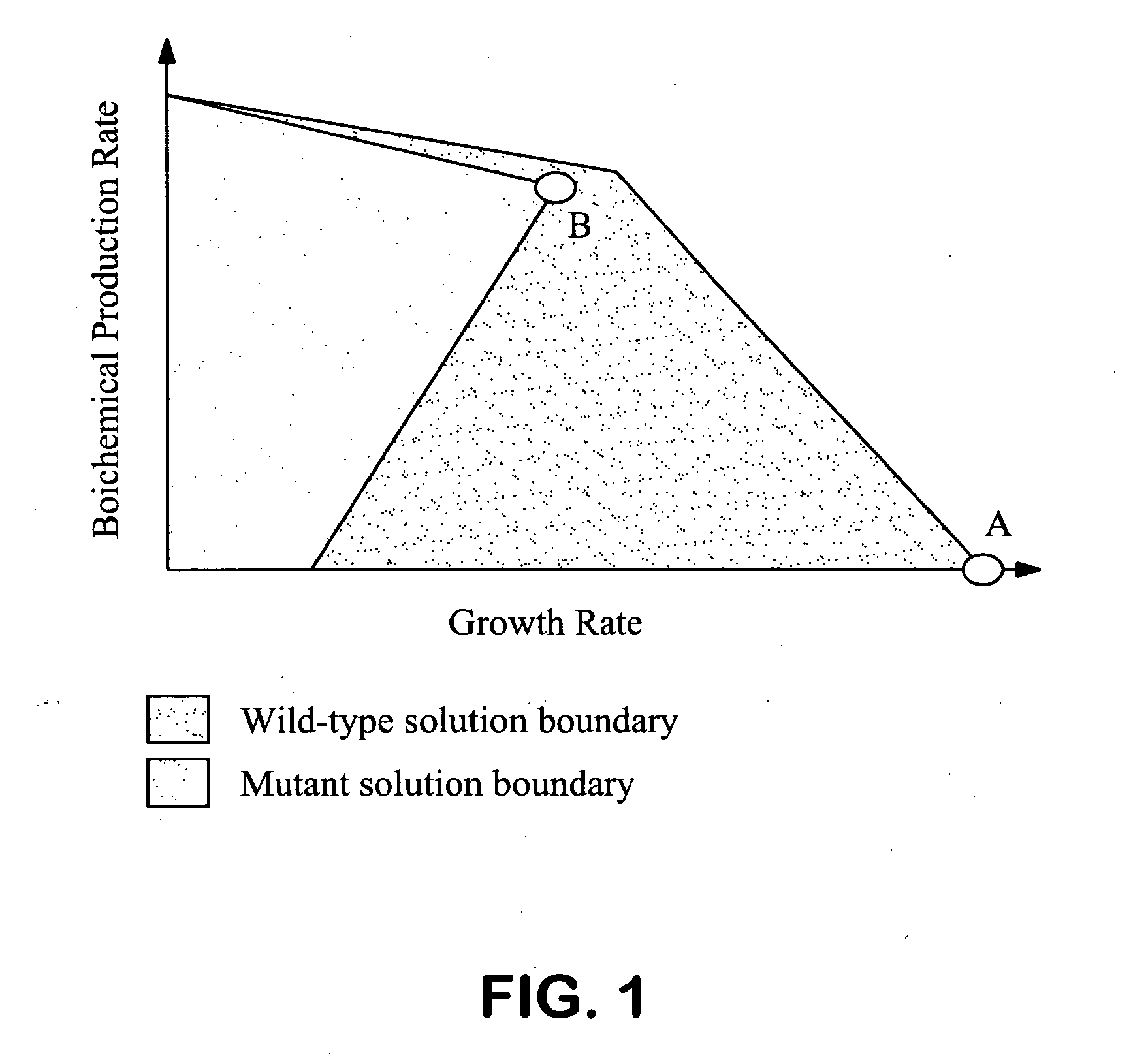
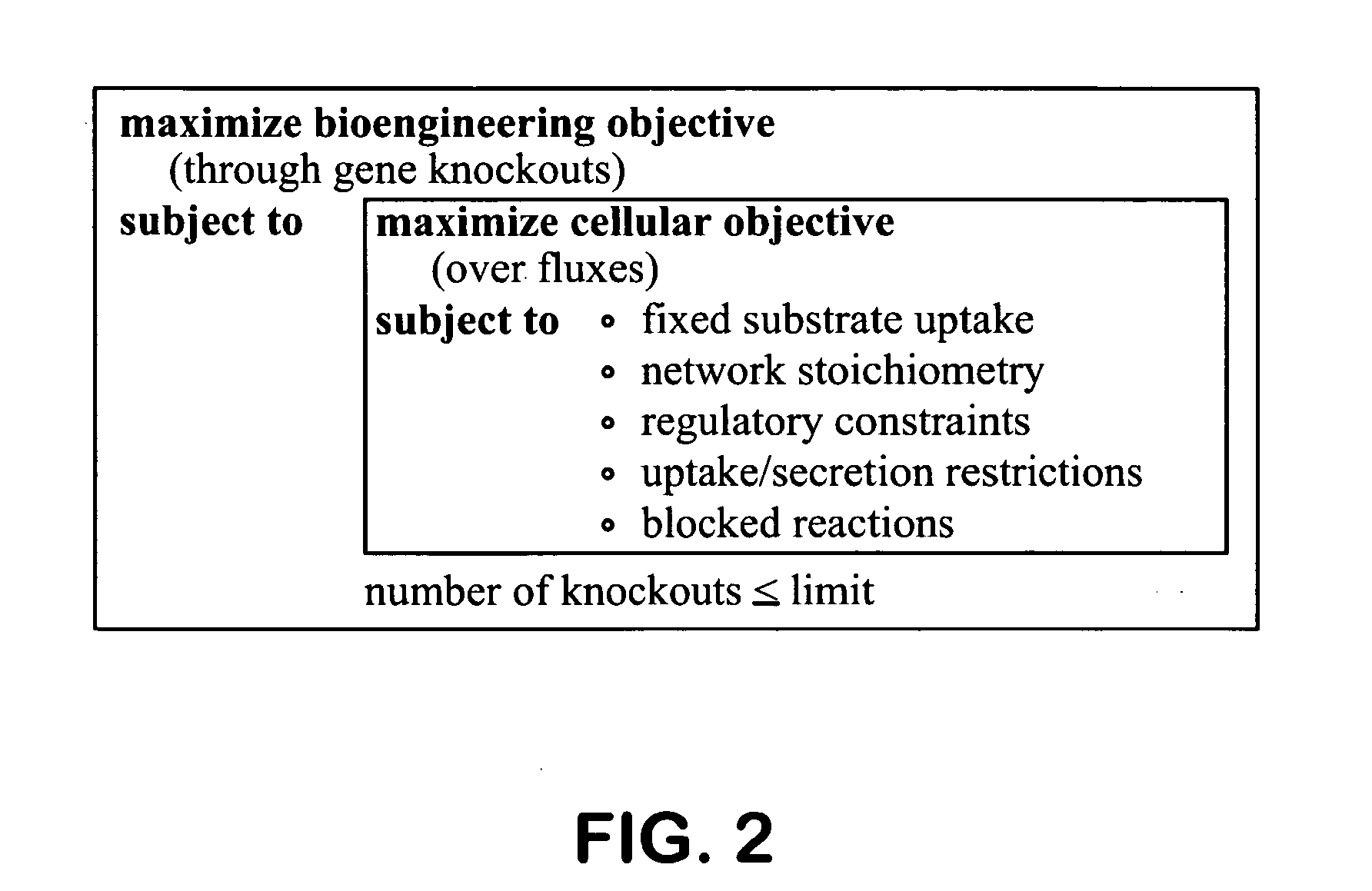
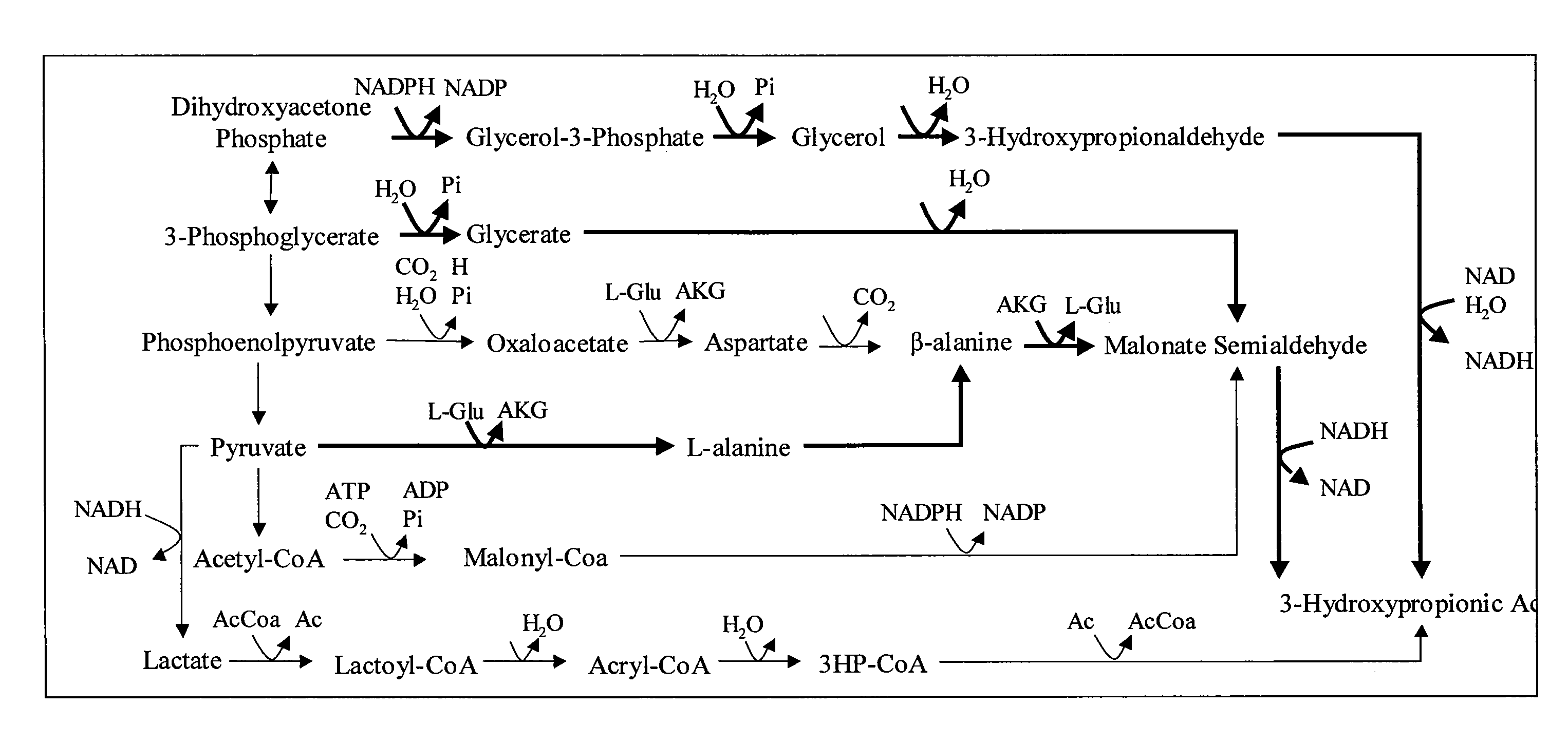
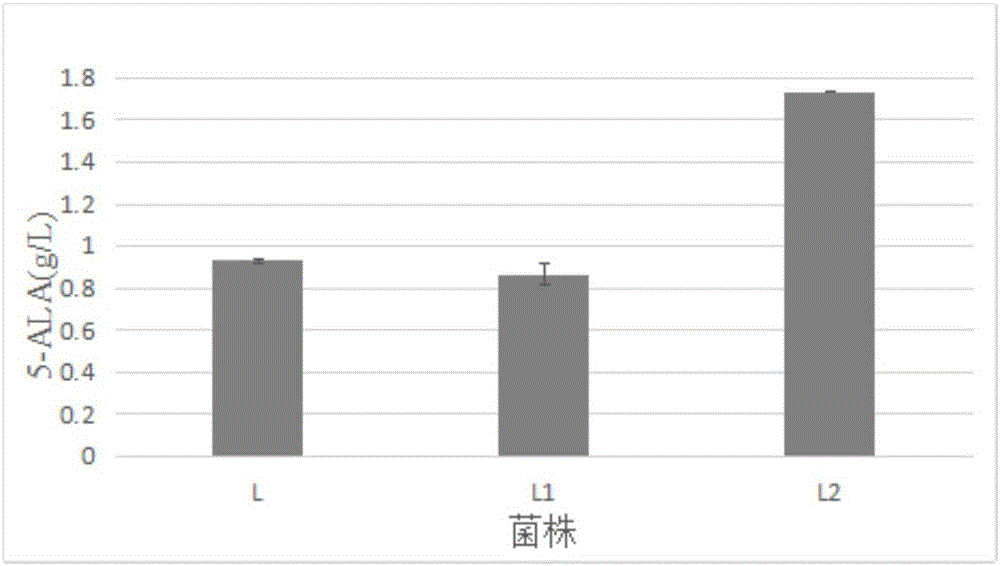
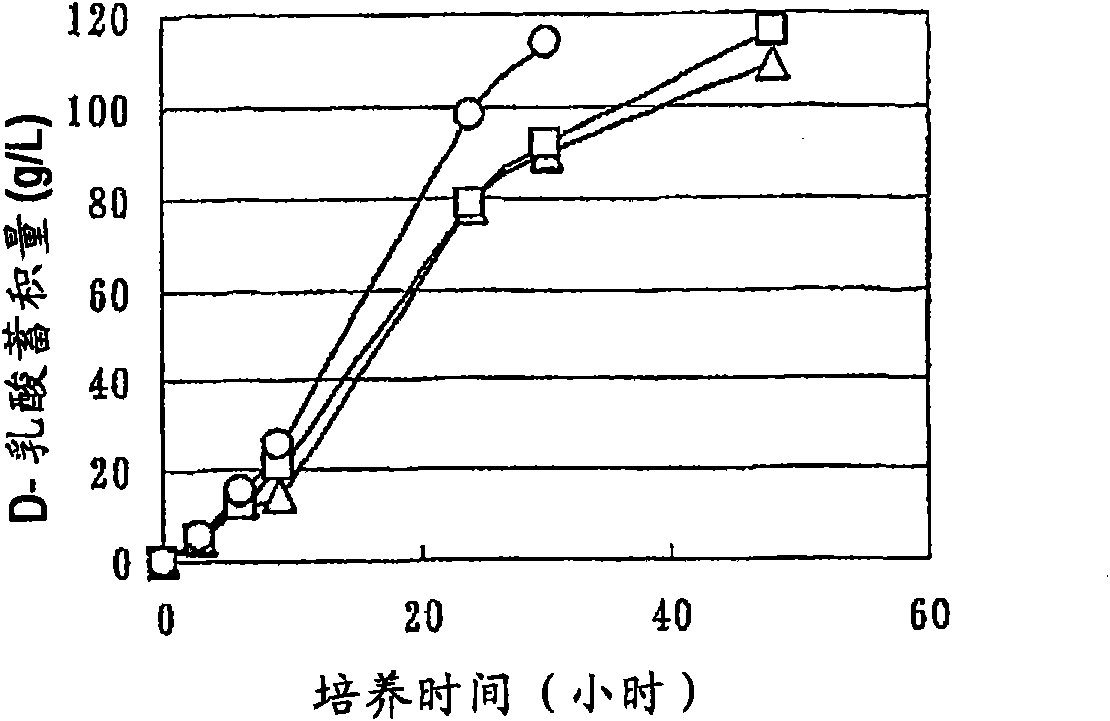
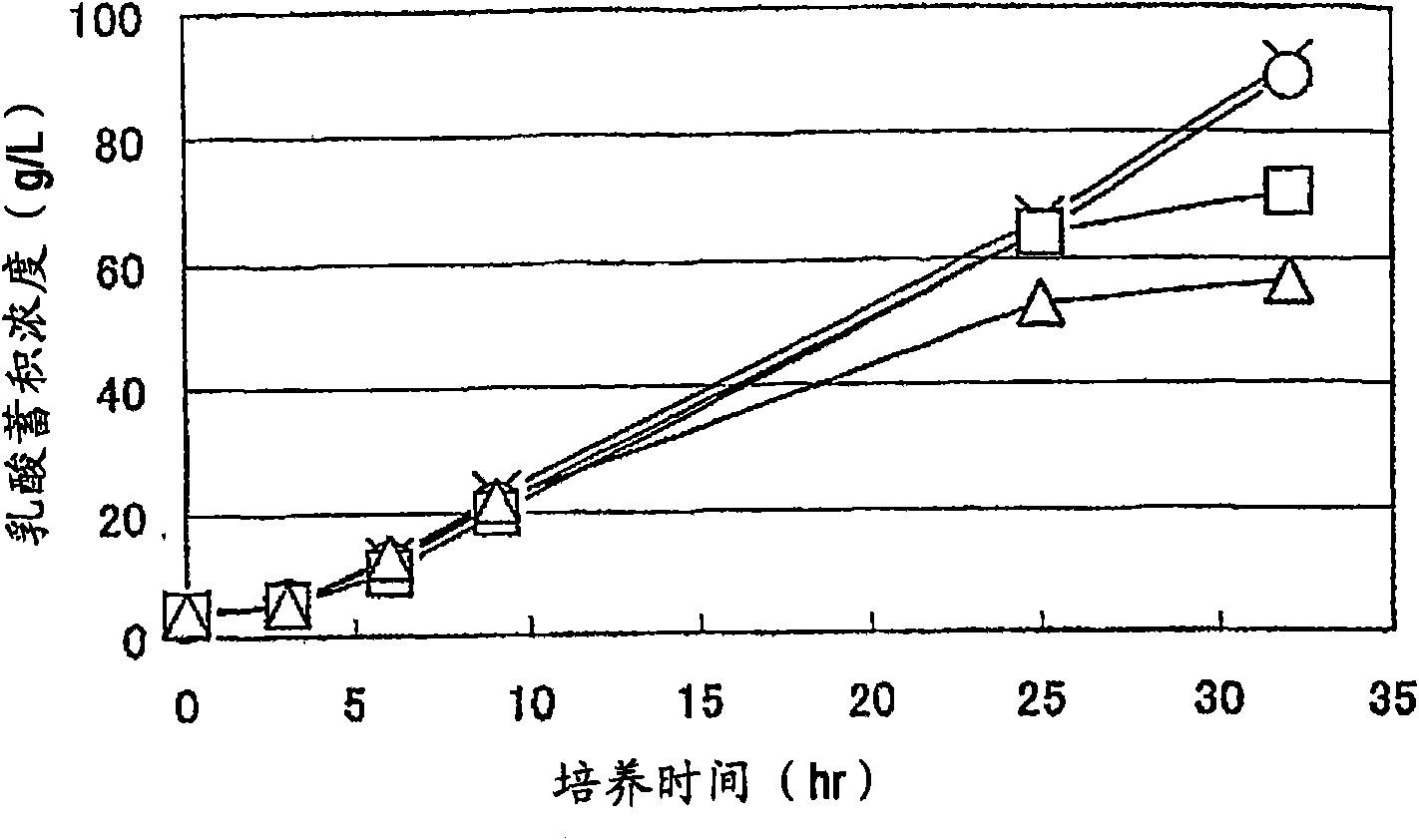
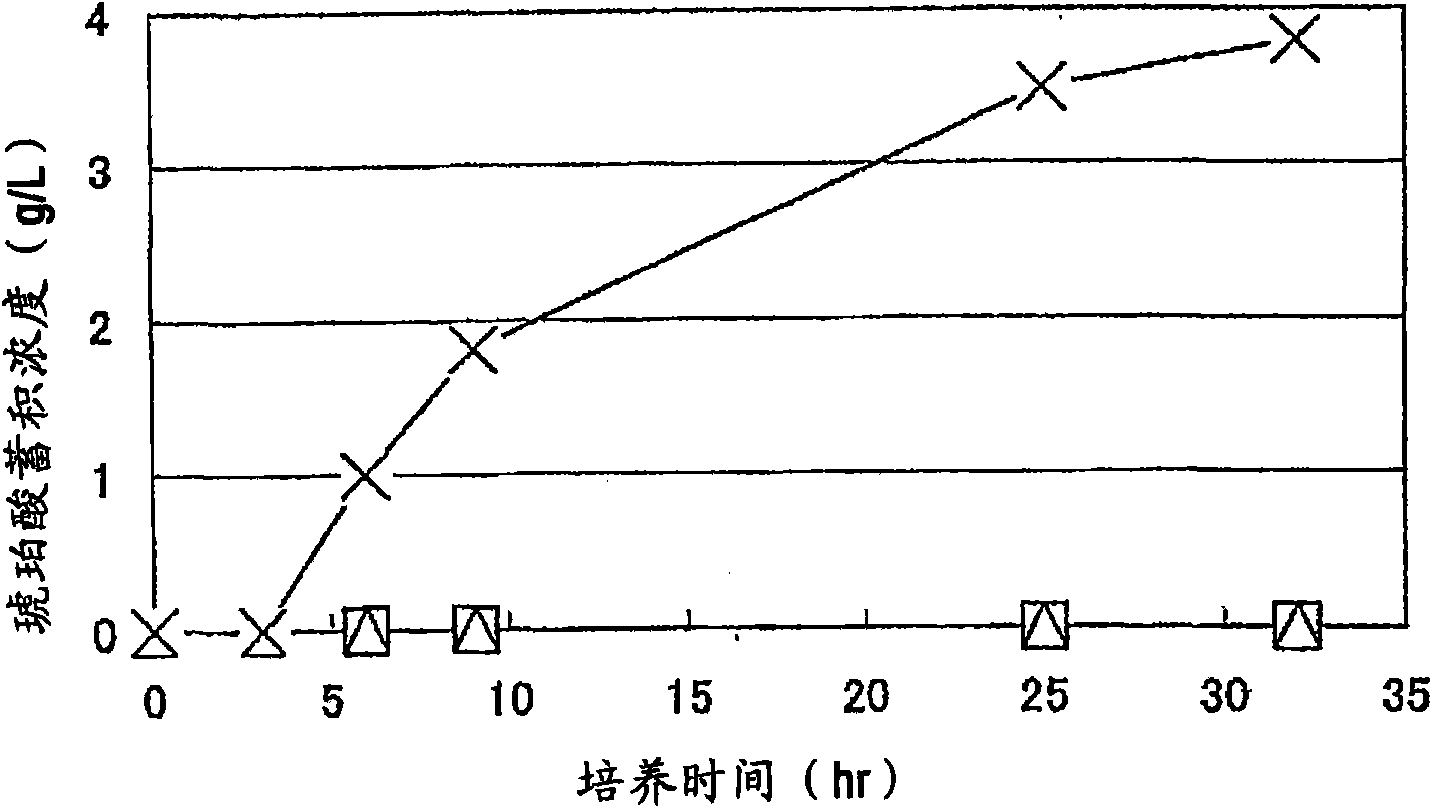
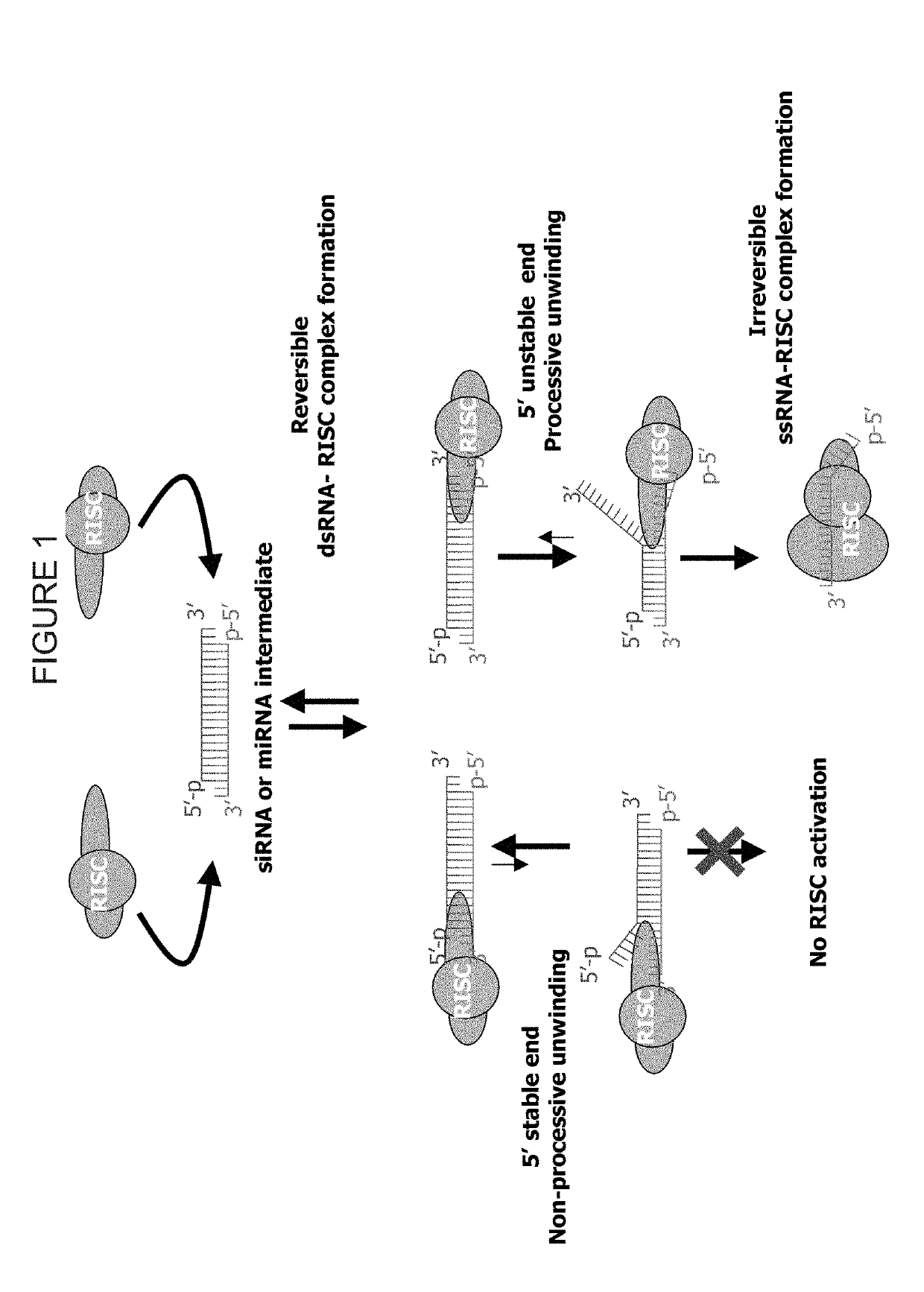
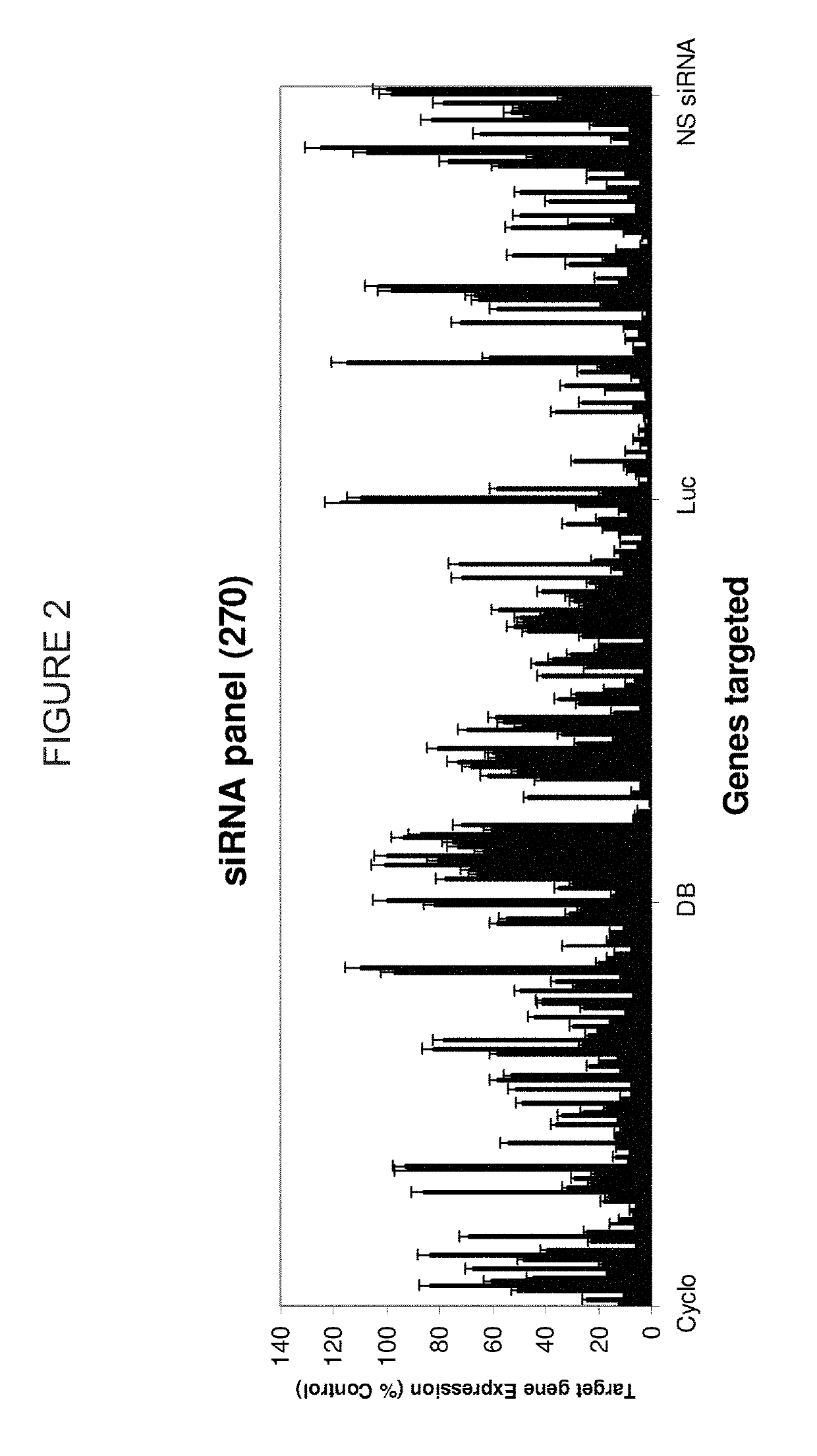
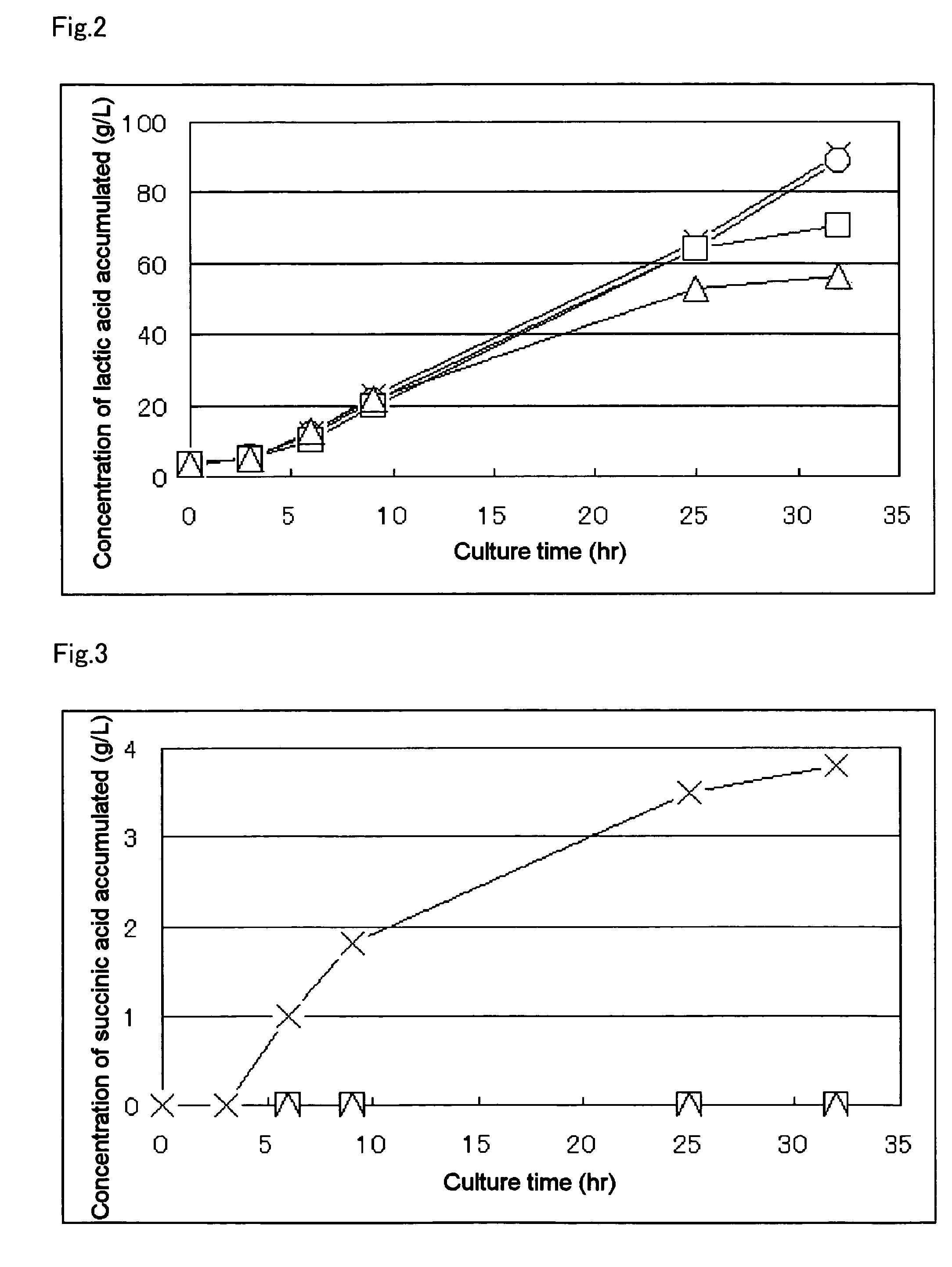
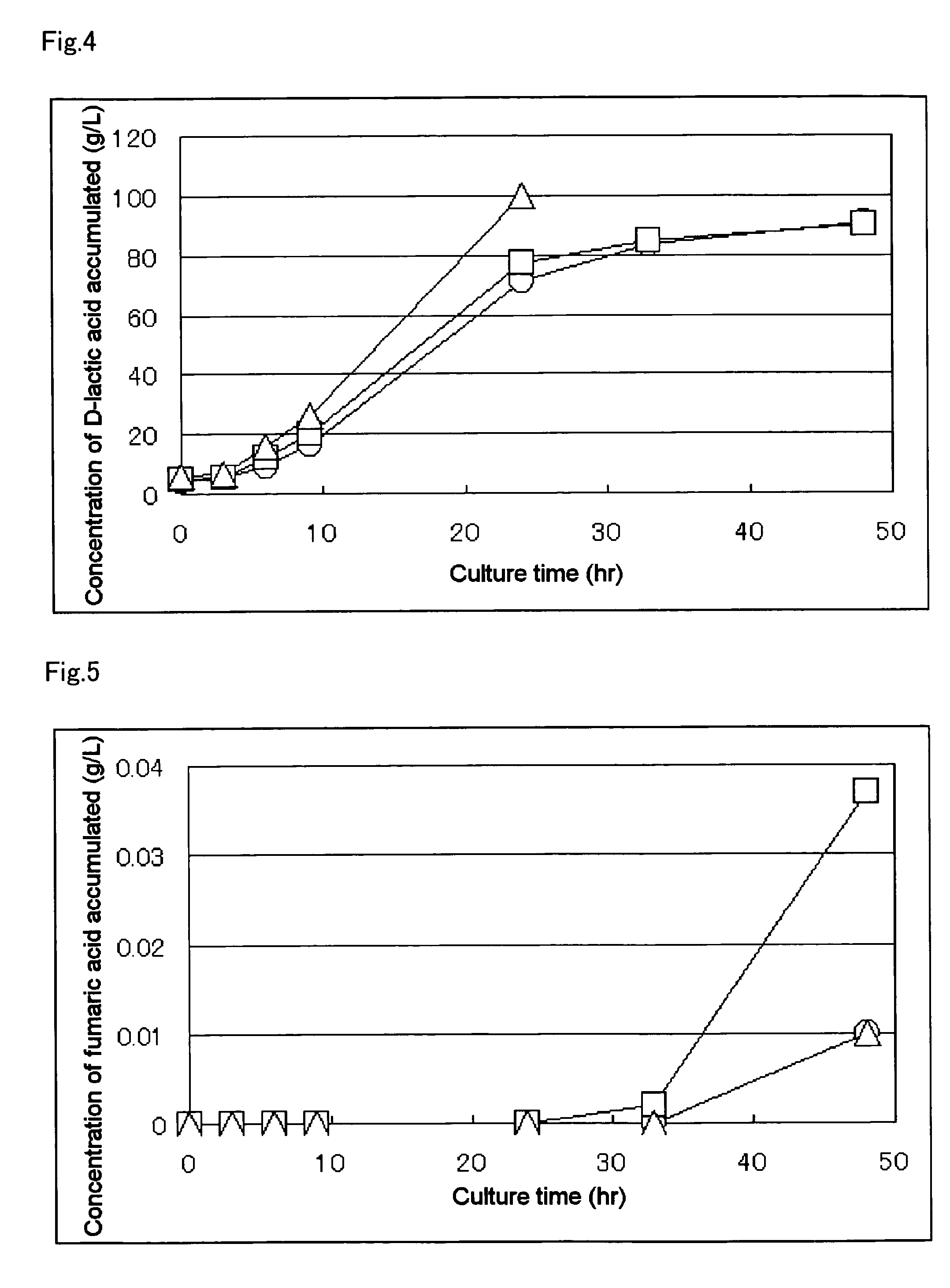
Patents
Literature
37 results about "Lactate Dehydrogenase A Gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Lactate dehydrogenase A (LDHA) is an enzyme which in humans is encoded by the LDHA gene. It is a monomer of lactate dehydrogenase, which exists as a tetramer. The other main subunit is lactate dehydrogenase B (LDHB).
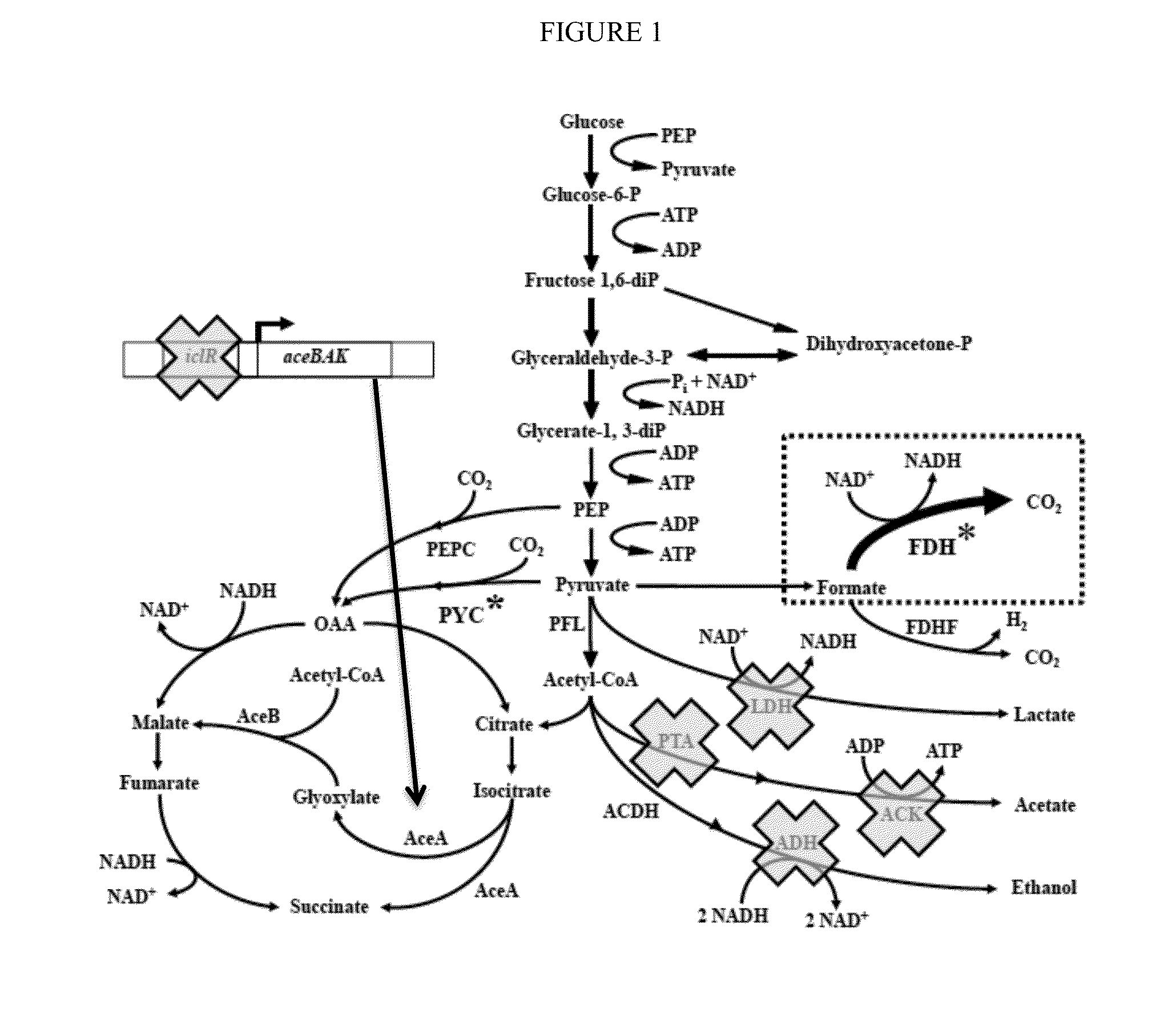
Methods and organisms for the growth-coupled production of succinate
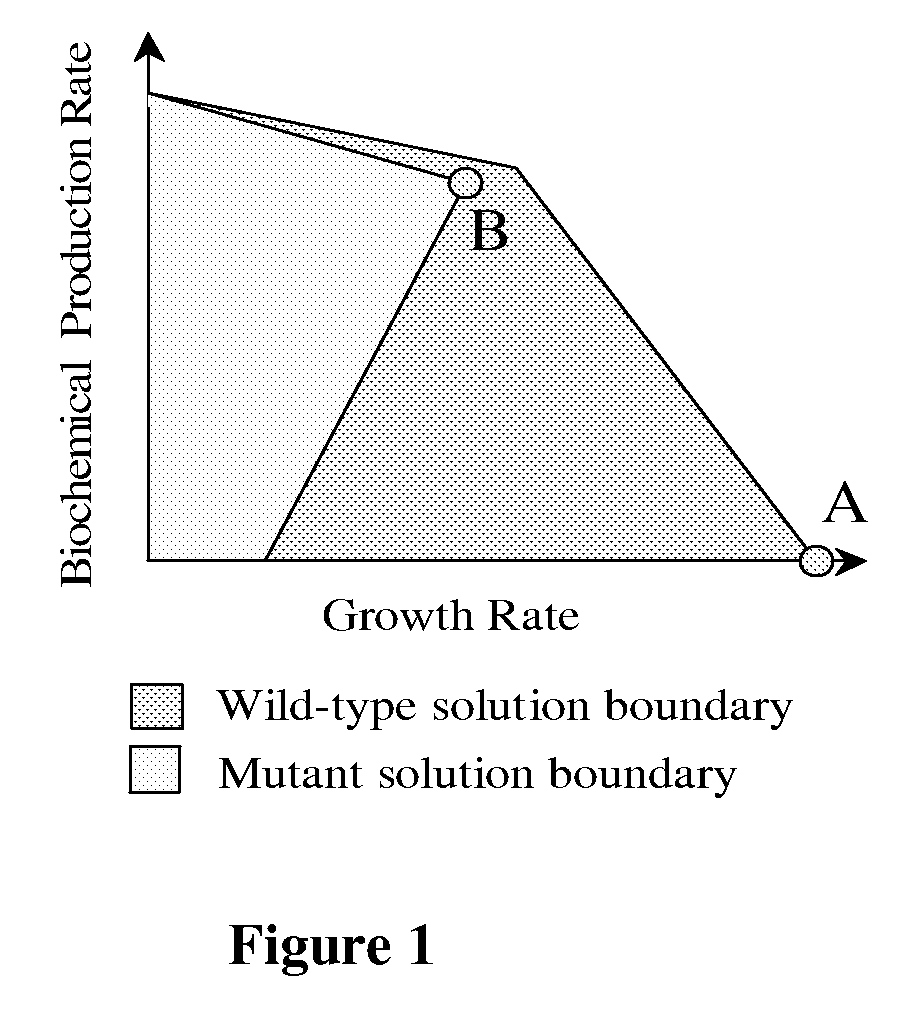
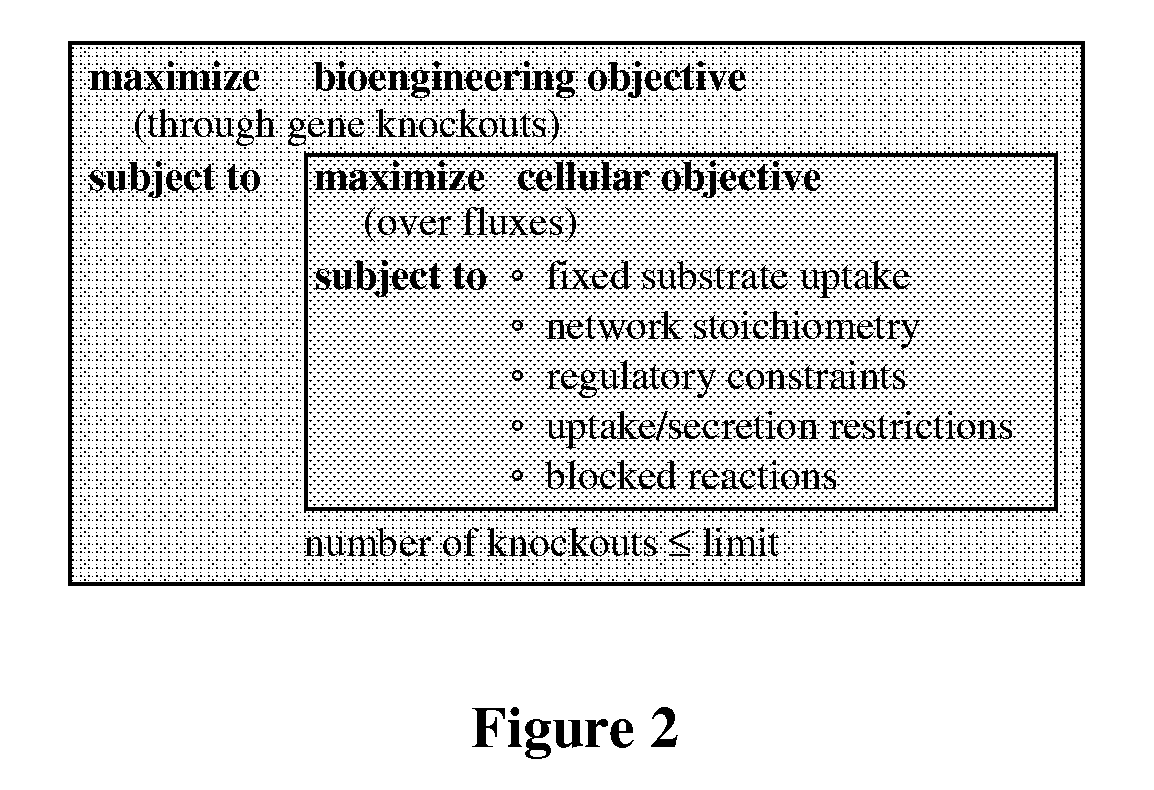
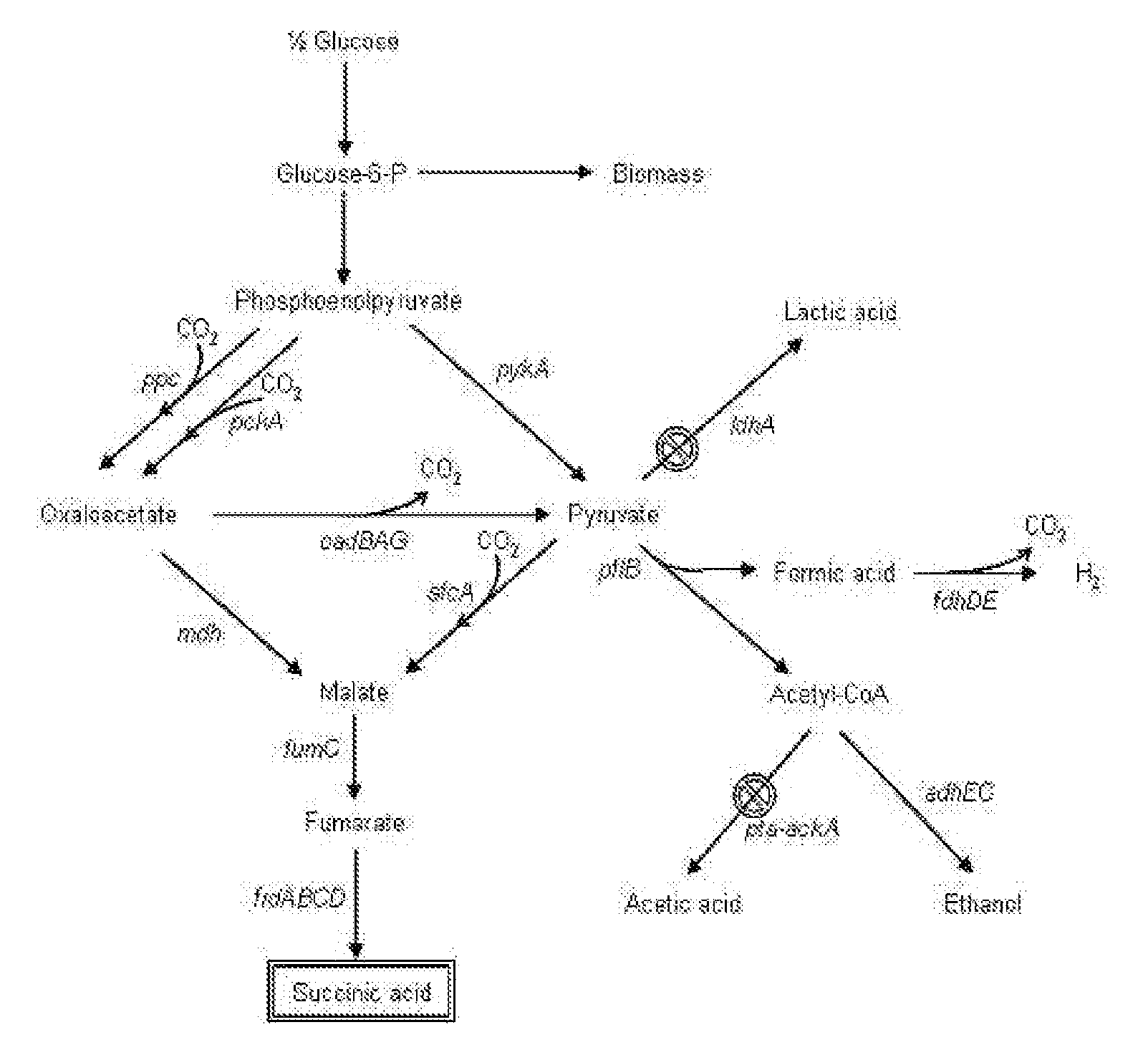
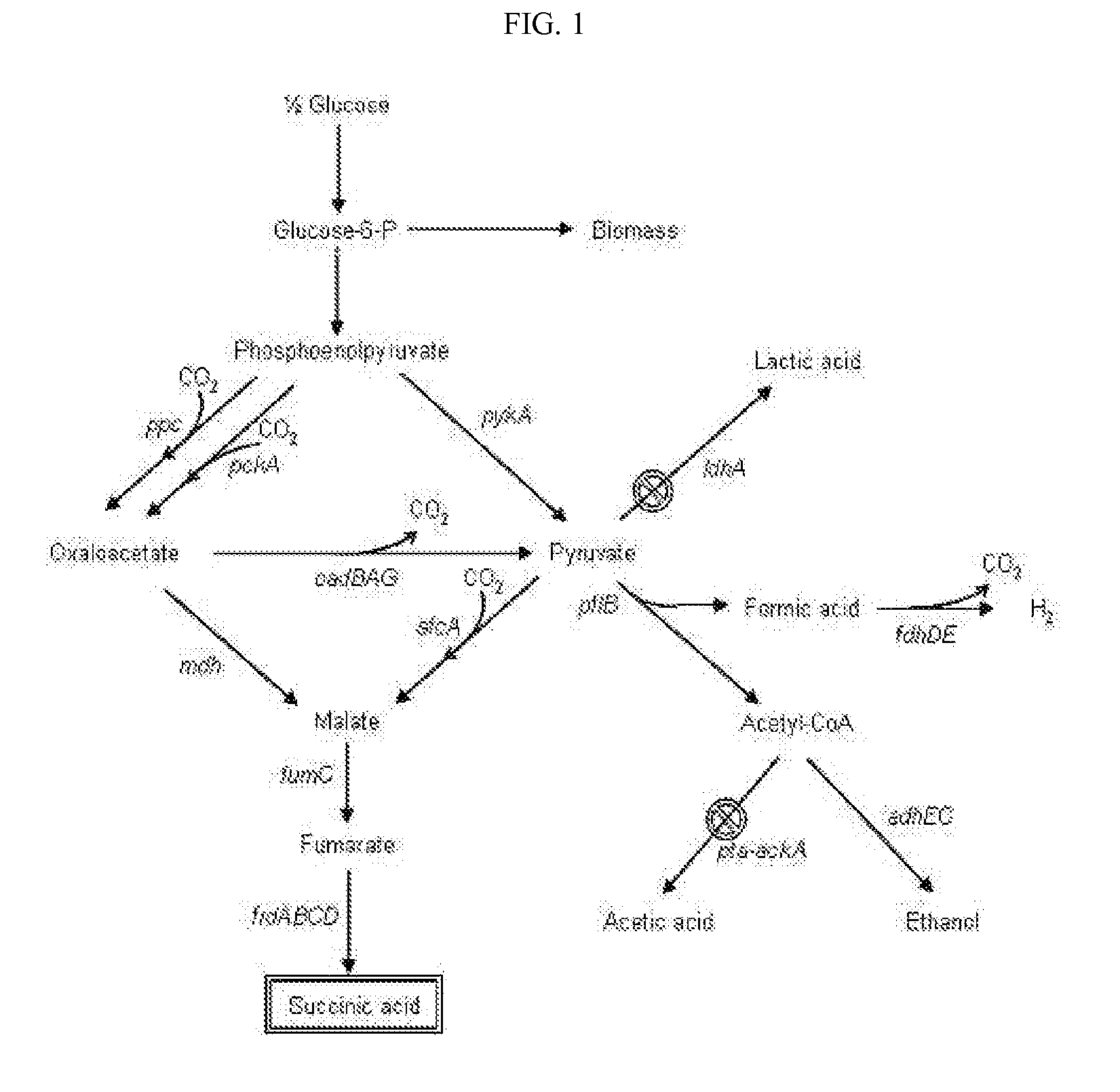
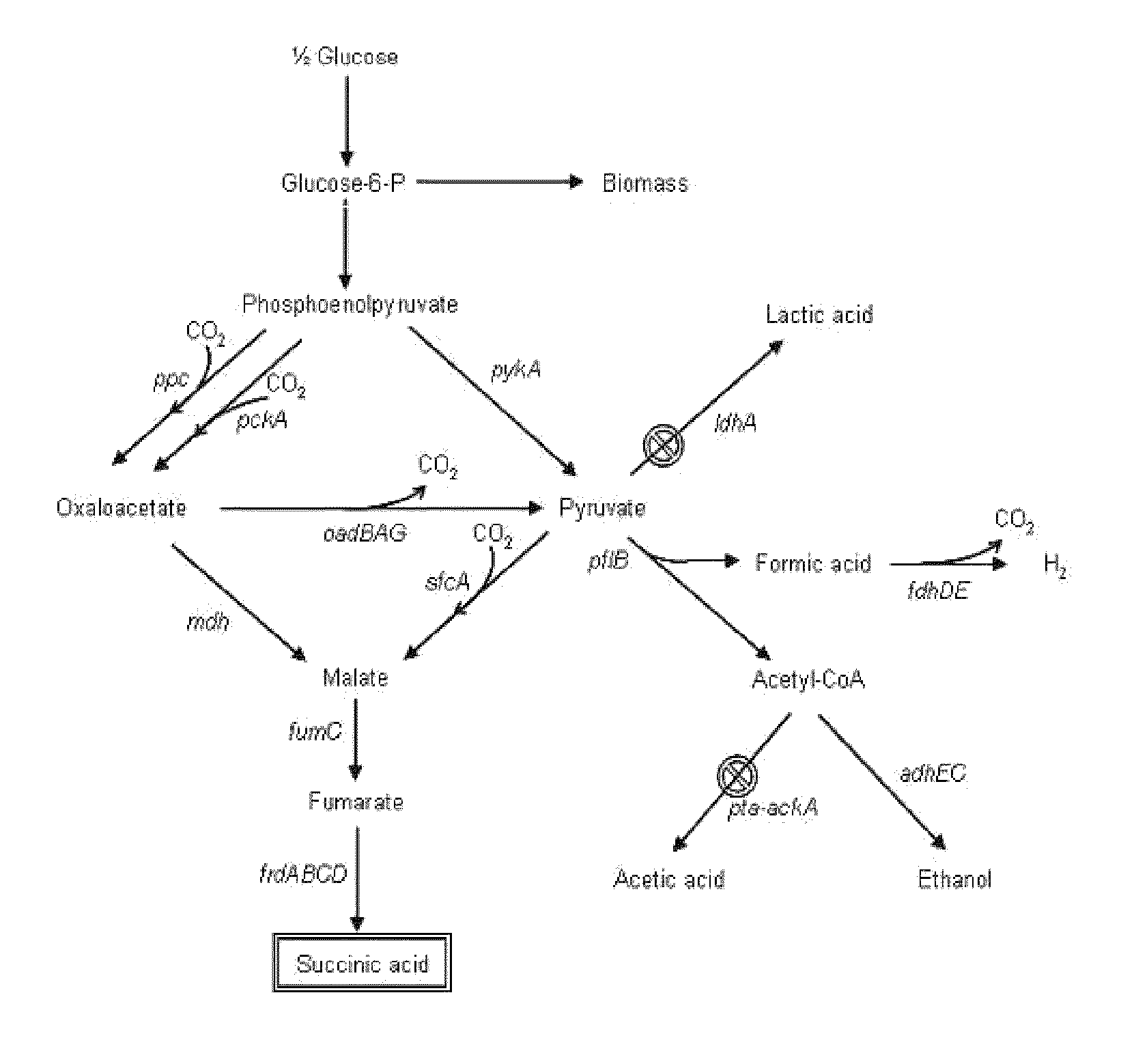
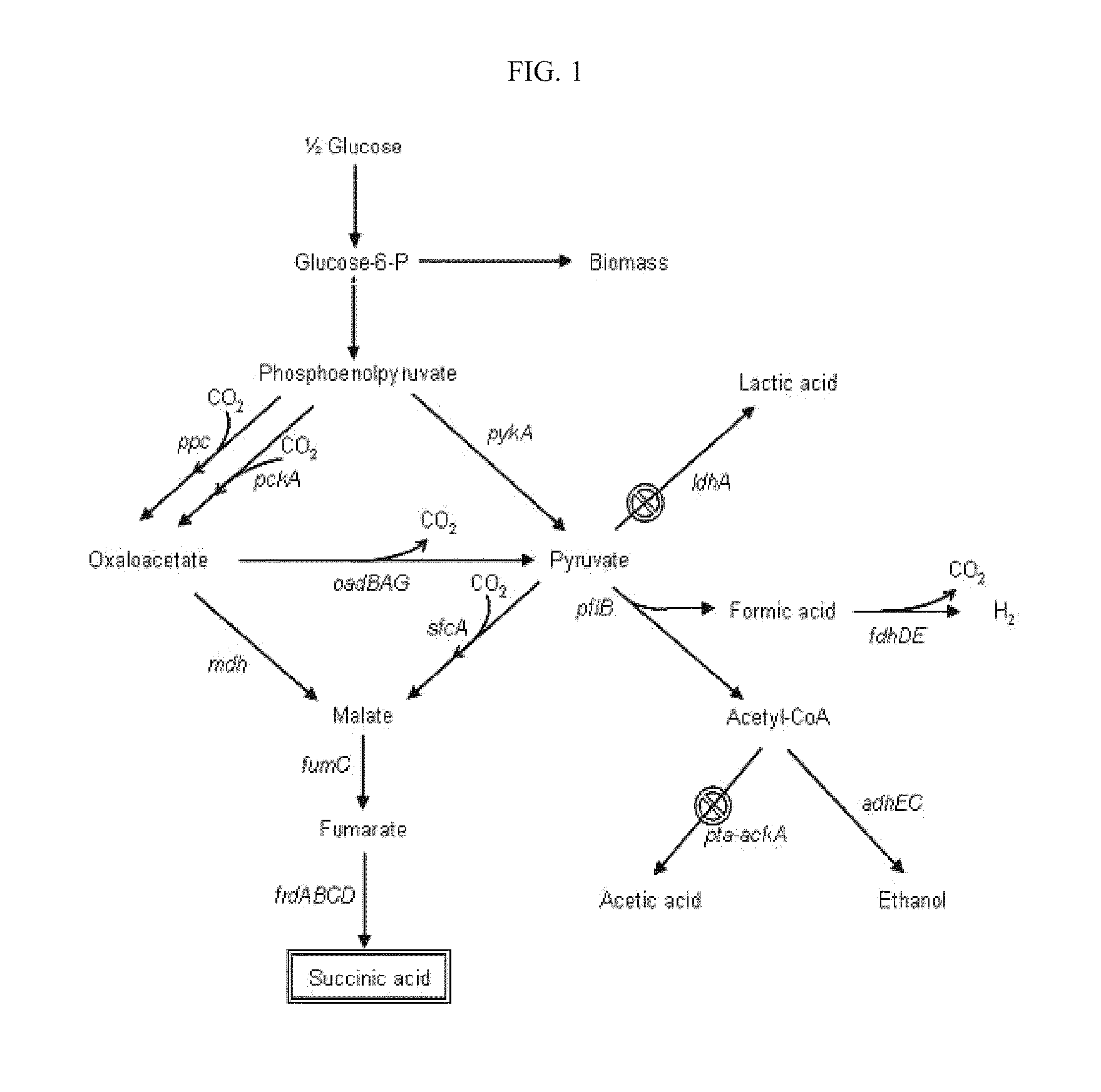
The invention provides a non-naturally occurring microorganism comprising one or more gene disruptions encoding an enzyme associated with growth-coupled production of succinate when an activity of the enzyme is reduced, whereby the one or more gene disruptions confers stable growth-coupled production of succinate onto the non- naturally occurring microorganism. Also provided is a non-naturally occurring microorganism comprising a set of metabolic modifications obligatory coupling succinate production to growth of the microorganism, the set of metabolic modifications comprising disruption of one or more genes selected from the set of genes comprising: (a) adhE, ldhA; (b) adhE, ldhA, acka-pta; (c) pfl, ldhA; (d) pfl, ldhA, adhE; (e) acka-pta, pykF, atpF, sdhA; (f) acka-pta, pykF, ptsG, or (g) acka-pta, pykF, ptsG, adhE, ldhA, or an ortholog thereof, wherein the microorganism exhibits stable growth-coupled production of succinate. Additionally provided is a non-naturally occurring microorganism having the genes encoding the metabolic modification (e) acka-pta, pykF, atpF, sdhA that further includes disruption of at least one gene selected from pyka, atpH, sdhB or dhaKLM; a non-naturally occurring microorganism having the genes encoding the metabolic modification (f) ackA-pta, pykF, ptsG that further includes disruption of at least one gene selected from pykA or dhaKLM, or a non-naturally occurring microorganism having the genes encoding the metabolic modification (g) ackA-pta, pykF, ptsG, adhE, ldhA that further includes disruption of at least one gene selected from pykA or dhaKLM. The disruptions can be complete gene disruptions and the non-naturally occurring organisms can include a variety of prokaryotic or eukaryotic microorganisms. A method of producing a non-naturally occurring microorganism having stable growth-coupled production of succinate also is provided. The method includes: (a) identifying in silico a set of metabolic modifications requiring succinate production during exponential growth, and (b) genetically modifying a microorganism to contain the set of metabolic modifications requiring succinate production.
Owner:GENOMATICA INC
Methods and Organisms for Growth-Coupled Production of 3-Hydroxypropionic Acid
The invention provides a non-naturally occurring microorganism having one or more gene disruptions, the one or more gene disruptions occurring in genes encoding an enzyme obligatory coupling 3-hydroxypropionic acid production to growth of the microorganism when the gene disruption reduces an activity of the enzyme, whereby the one or more gene disruptions confers stable growth-coupled production of 3-hydroxypropionic acid onto the non-naturally occurring microorganism. Also provided is a non-naturally occurring microorganism comprising a set of metabolic modifications obligatory coupling 3-hydroxypropionic acid production to growth of the microorganism, the set of metabolic modifications having disruption of one or more genes including: (a) the set of genes selected from: (1) adhE, ldhA, pta-ackA; (2) adhE, ldhA, frdABCD; (3) adhE, ldhA, frdABCD, ptsG; (4) adhE, ldhA, frdABCD, pntAB; (5) adhE, ldhA, fumA, fumB, fumC; (6) adhE, ldhA, fumA, fumB, fumC, pntAB; (7) pflAB, ldhA, or (8) adhE, ldhA, pgi in a microorganism utilizing an anaerobic β-alanine 3-HP precursor pathway; (b) the set of genes selected from: (1) tpi, zwf; (2) tpi, ybhE; (3) tpi, gnd; (4) fpb, gapA; (5) pgi, edd, or (6) pgi, eda in a microorganism utilizing an aerobic glycerol 3-HP precursor pathway; (c) the set of genes selected from: (1) eno; (2) yibO; (3) eno, atpH, or other atp subunit, or (4) yibO, atpH, or other atp subunit, in a microorganism utilizing a glycerate 3-HP precursor pathway, or an ortholog thereof, wherein the microorganism exhibits stable growth-coupled production of 3-hydroxypropionic acid. The disruptions can be complete gene disruptions and the non-naturally occurring organisms can include a variety of prokaryotic or eukaryotic microorganisms. A method of producing a non-naturally occurring microorganism having stable growth-coupled production of 3-hydroxypropionic acid is further provided. The method includes: (a) identifying in silico a set of metabolic modifications requiring 3-hydroxypropionic acid production during exponential growth, and (b) genetically modifying a microorganism to contain the set of metabolic modifications requiring 3-hydroxypropionic acid production.
Owner:GENOMATICA INC
Simultaneous anaerobic production of isoamyl acetate and succinic acid
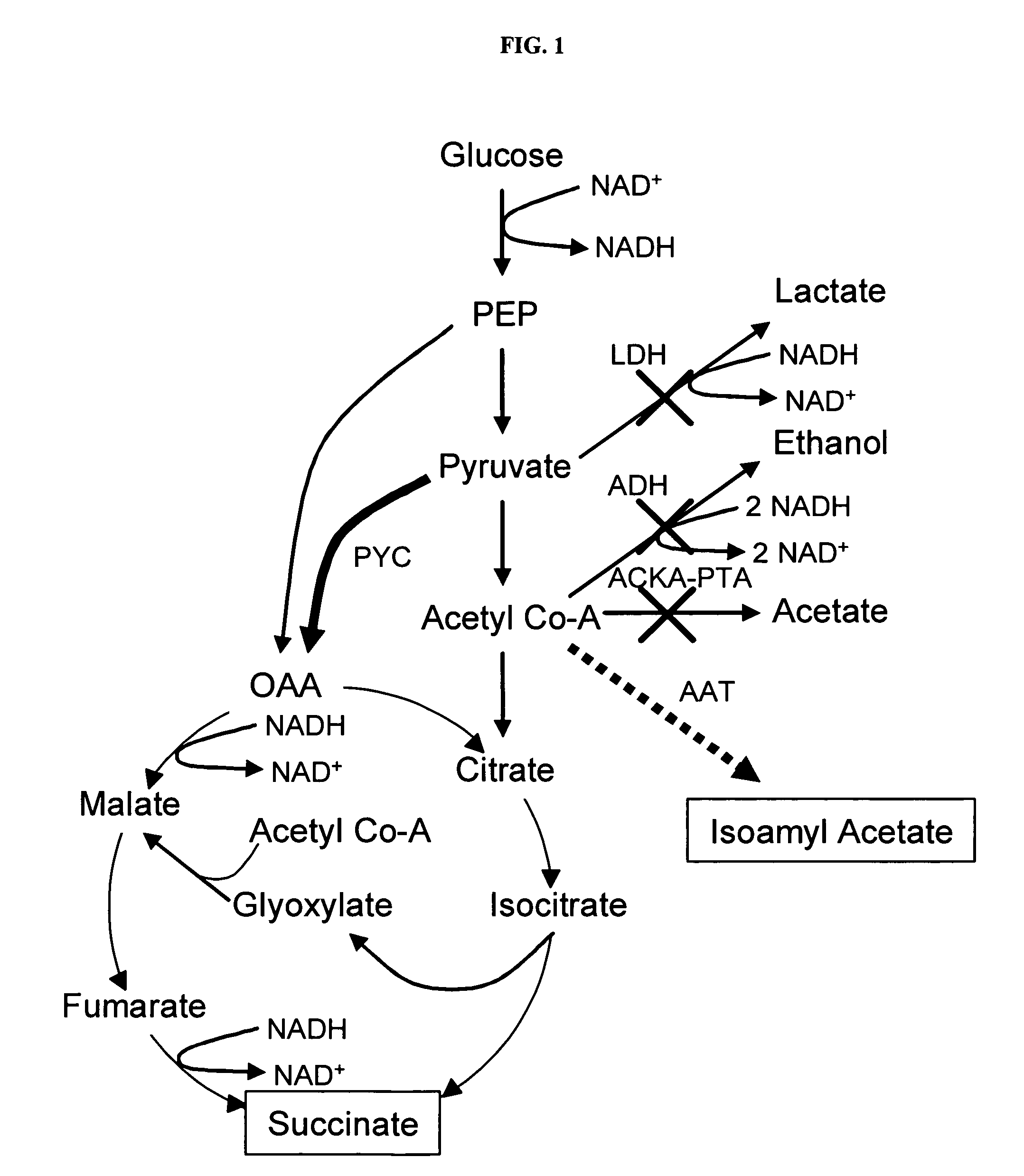
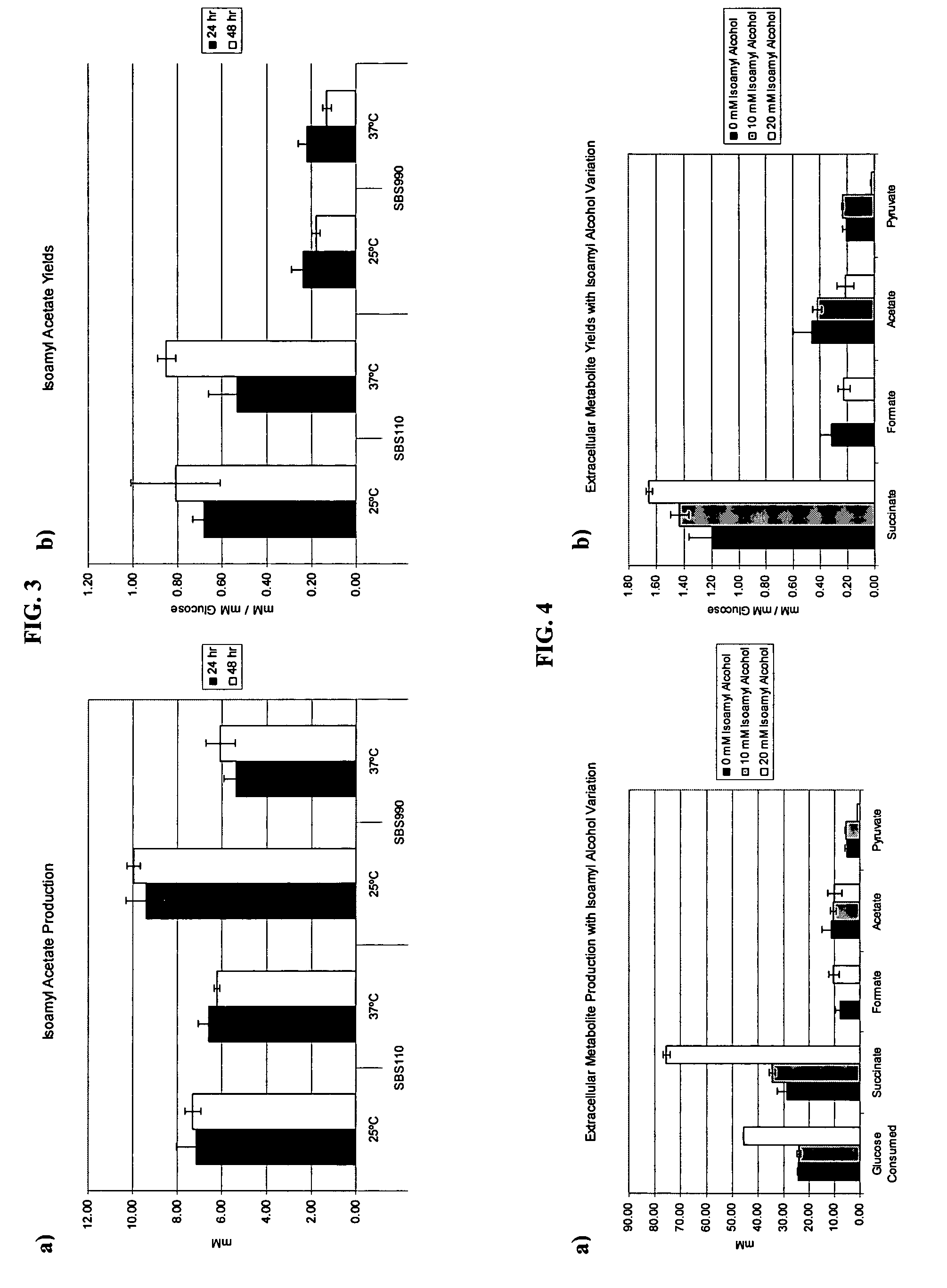
In vivo method of producing esters from acetyle coA, such as isoamyl acetate and succinate, has been developed by producing null mutants in pathways that use acetyl coA and by overexpressing products that use NADH and in order to maintain the proper redox balance between NADH and NAD+. The method is exemplified with null mutations in ldhA, adhE, ackA-pta and overexpression of pyruvate carboxylase and alcohol acetyltransferase. This strain produces higher levels of both isoamyl acetate and succinate.
Owner:RICE UNIV
Novel rumen bacteria variants and process for preparing succinic acid employing the same
ActiveUS20070054387A1High yieldBacteriaSugar derivativesHigh concentrationPhosphoenolpyruvate carboxylase
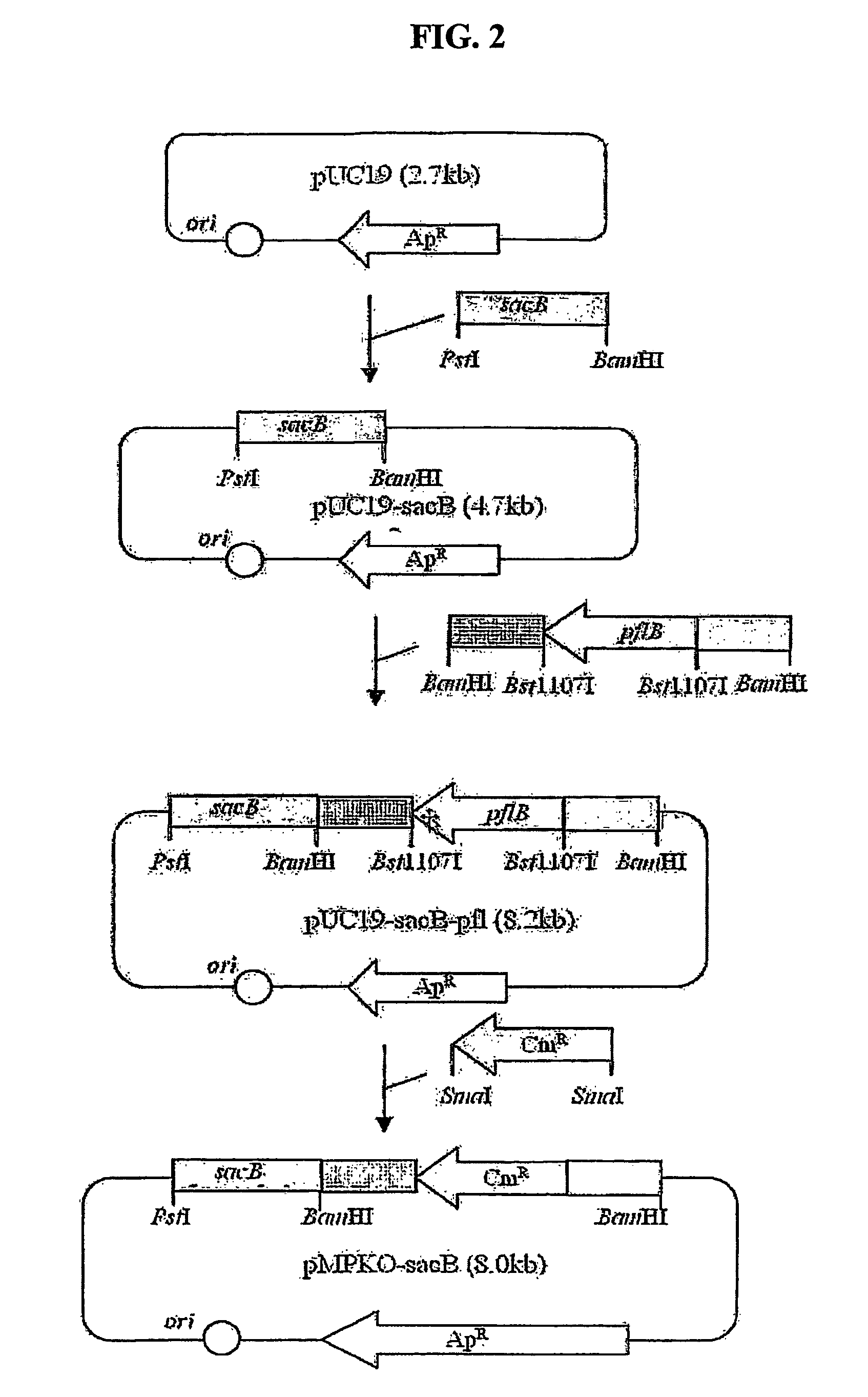
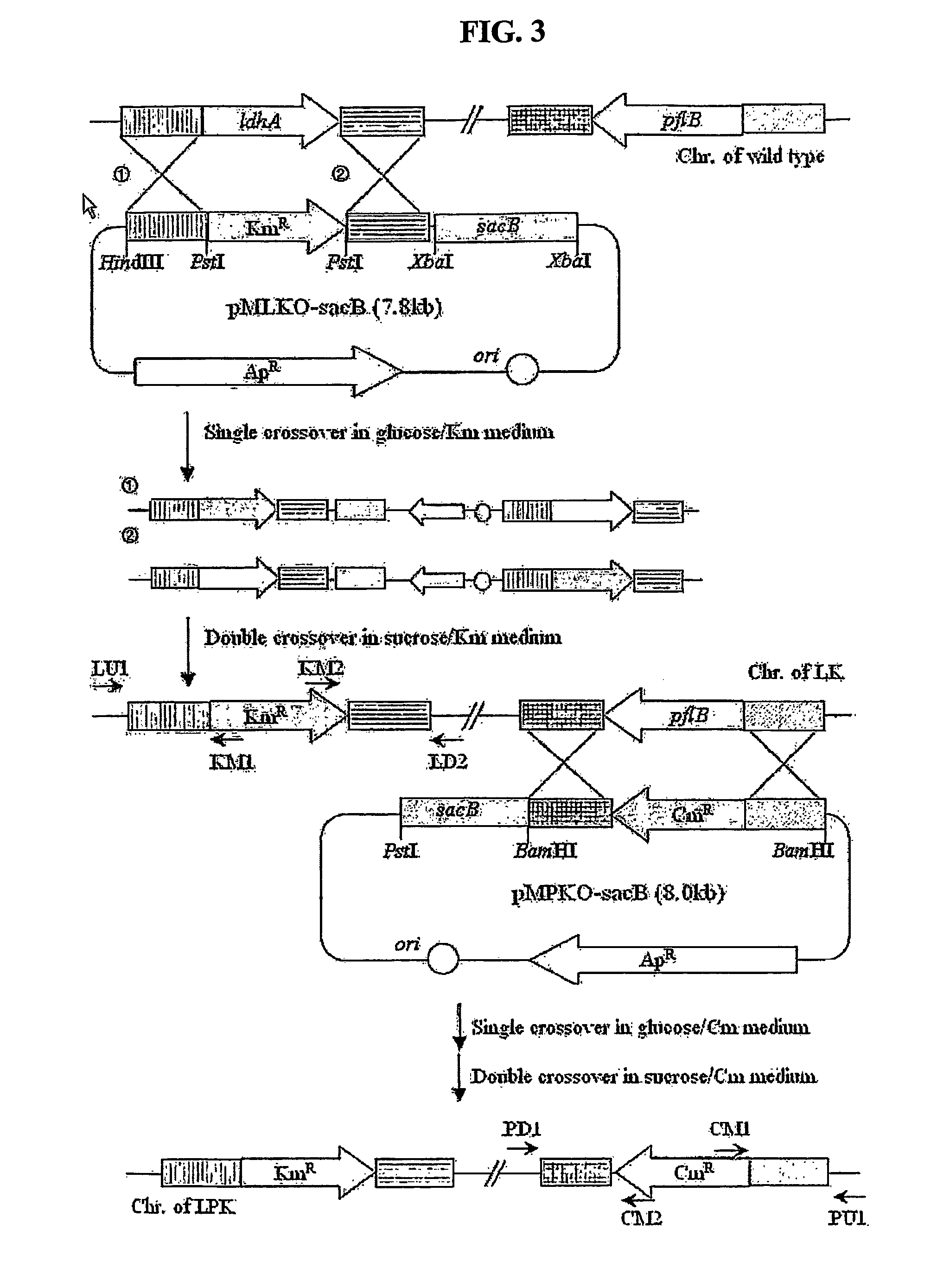
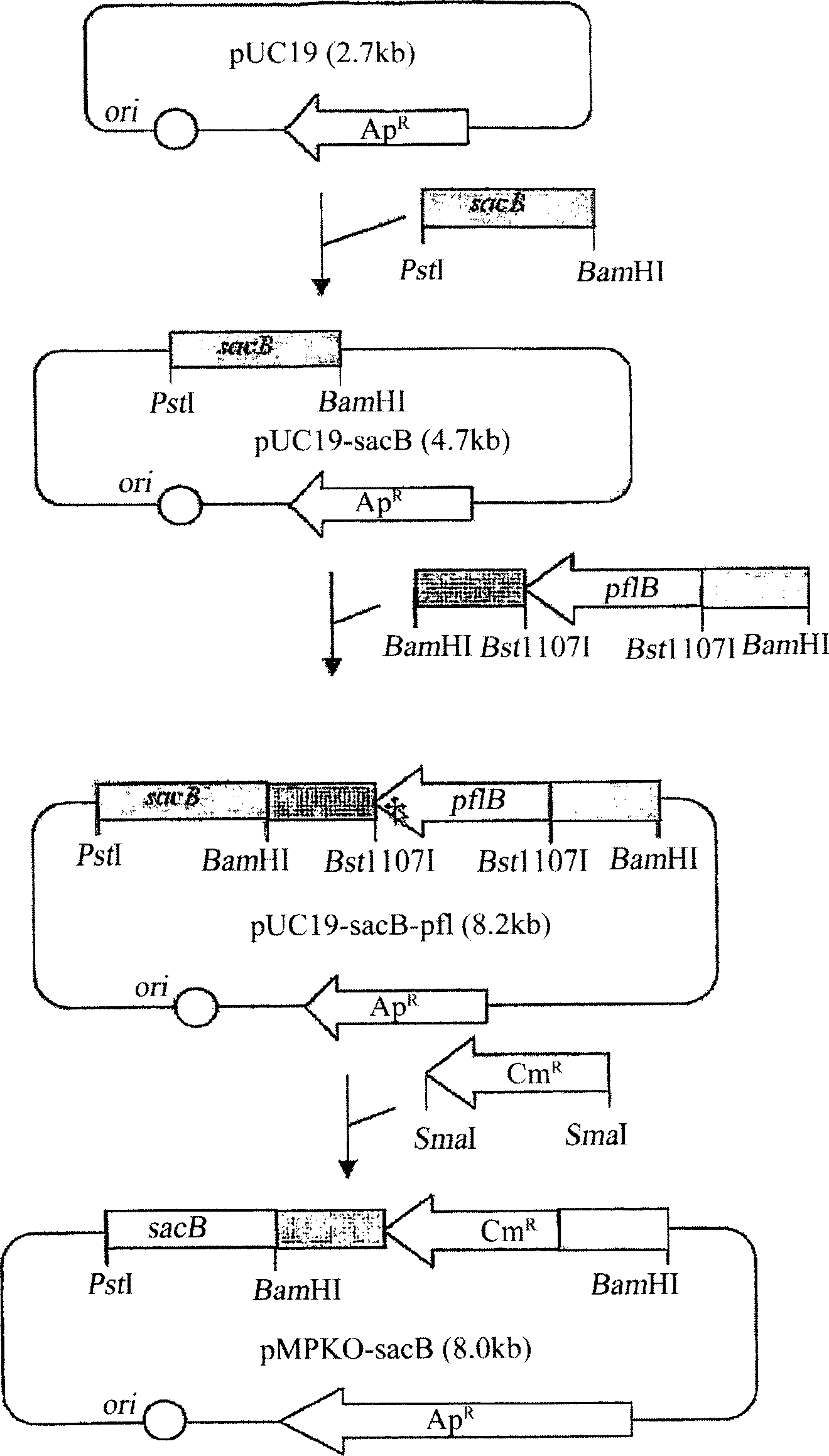
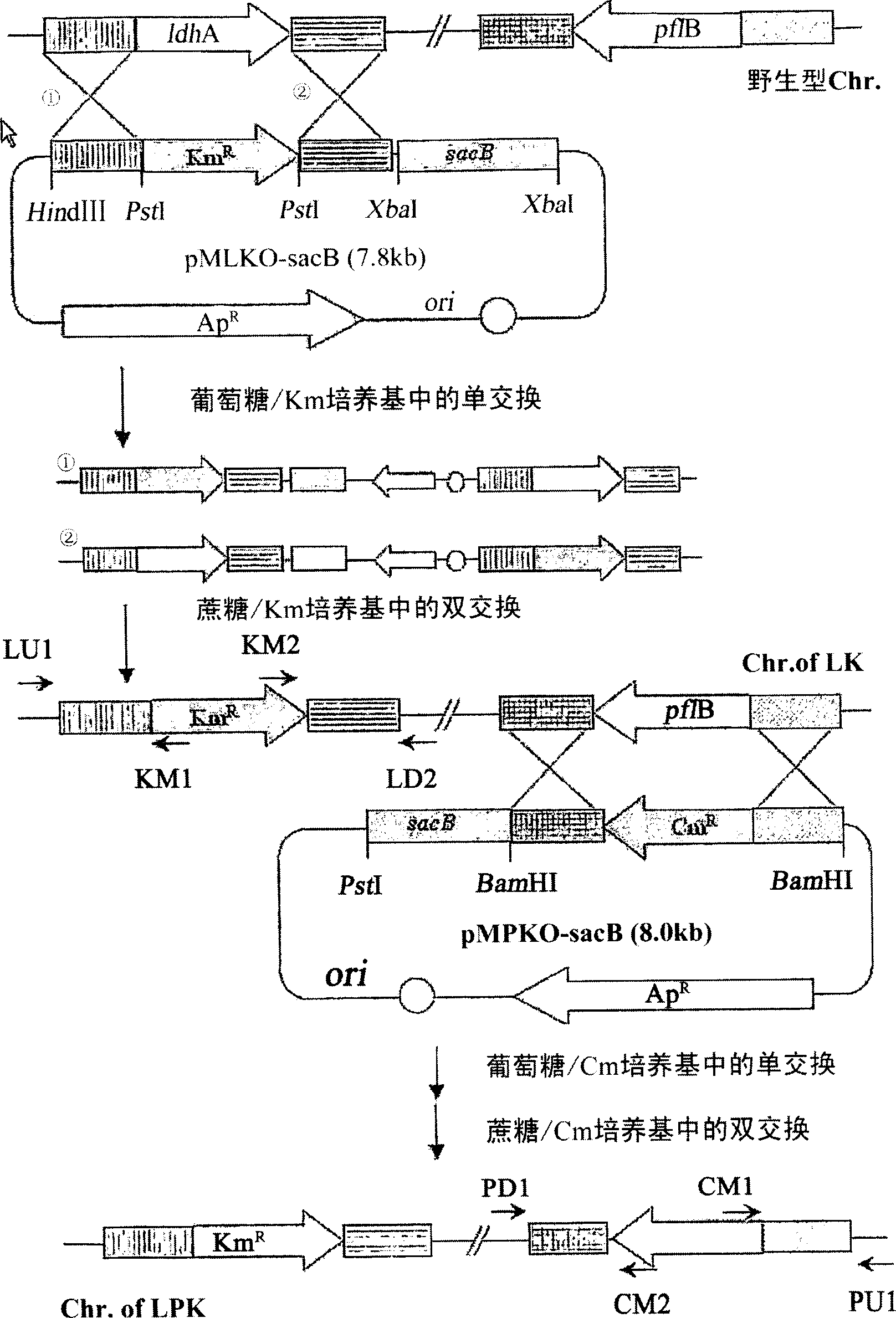
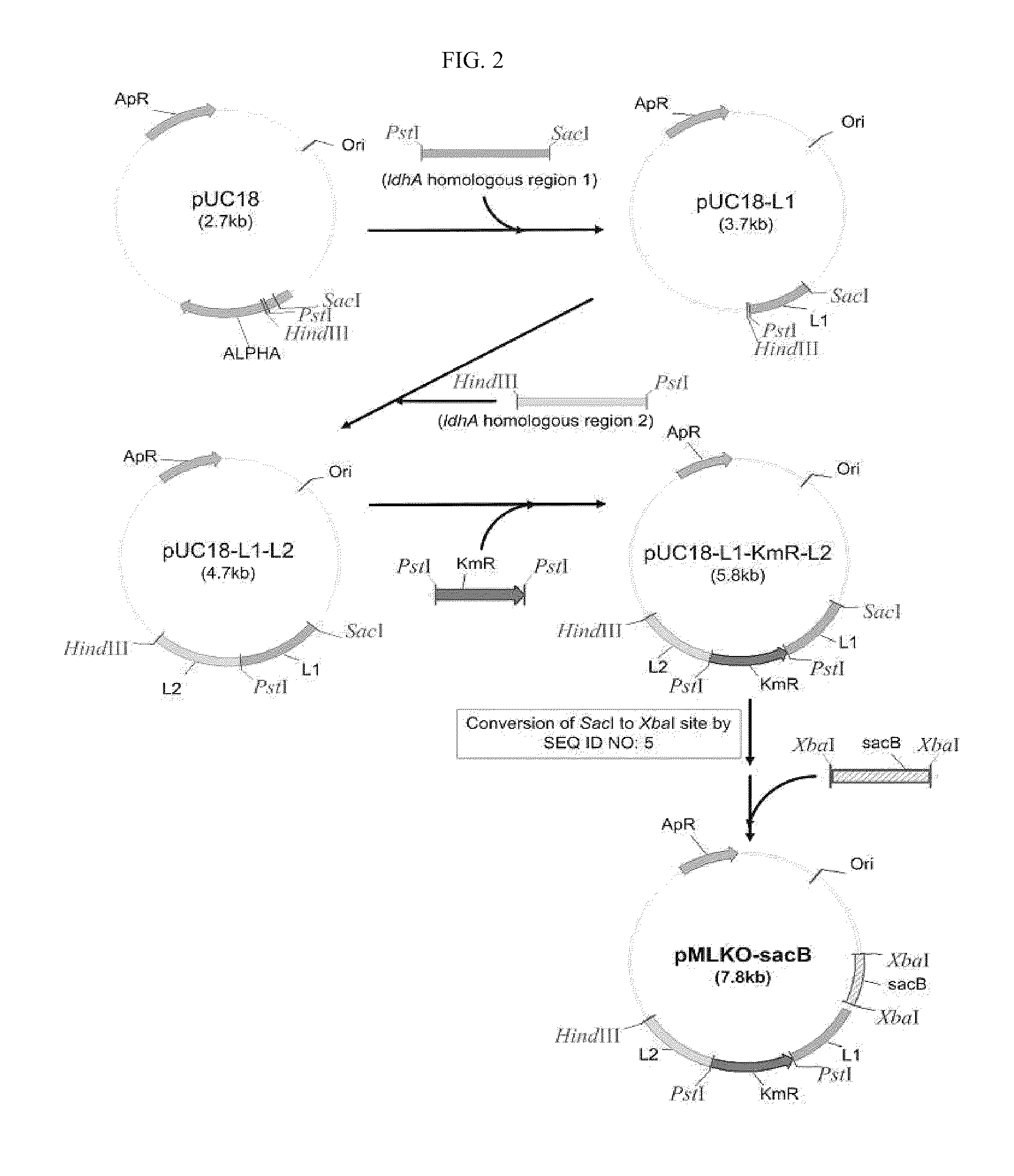
The present invention relates to novel rumen bacterial mutants resulted from the disruption of a lactate dehydrogenase gene (ldhA) and a pyruvate formate-lyase gene (pfl) (which are involved in the production of lactic acid, formic acid and acetic acid) from rumen bacteria; a novel bacterial mutant (Mannheimia sp. LPK7) having disruptions of a lactate dehydrogenase gene (ldhA), a pyruvate formate-lyase gene (pfl), a phosphotransacetylase gene (pta), and a acetate kinase gene (ackA); a novel bacterial mutant (Mannheimia sp. LPK4) having disruptions of a lactate dehydrogenase gene (ldhA), a pyruvate formate-lyase gene (pfl) and a phosphoenolpyruvate carboxylase gene (ppc) involved in the immobilization of CO2 in a metabolic pathway of producing succinic acid; and a method for producing succinic acid, which is characterized by the culture of the above mutants in anaerobic conditions. The inventive bacterial mutants have the property of producing succinic acid at high concentration while producing little or no organic acids, as compared to the prior wild-type strains of producing various organic acids. Thus, the inventive bacterial mutants are useful as strains for the industrial production of succinic acid.
Owner:KOREA ADVANCED INST OF SCI & TECH
High-yield succinic acid strain and its application
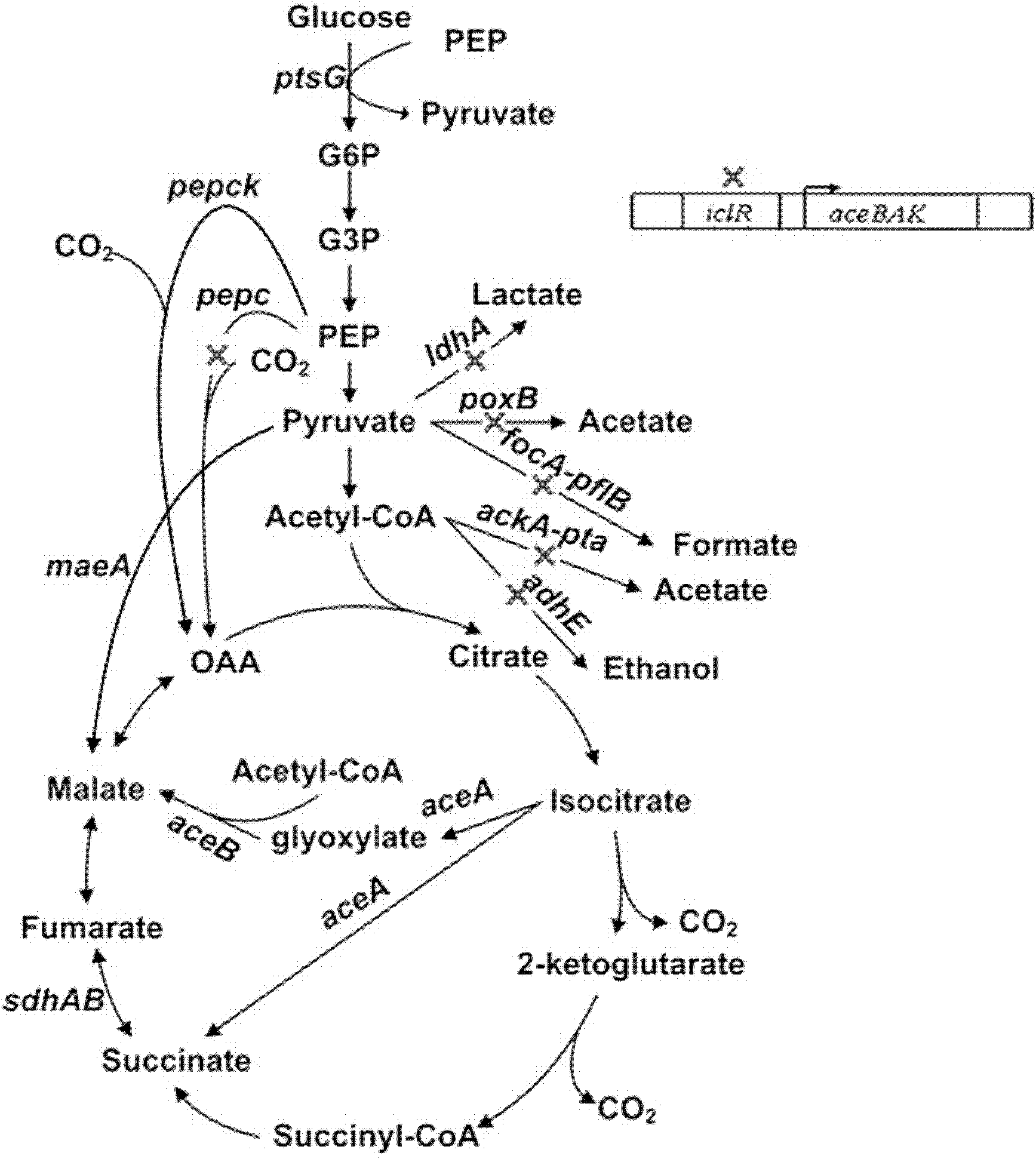
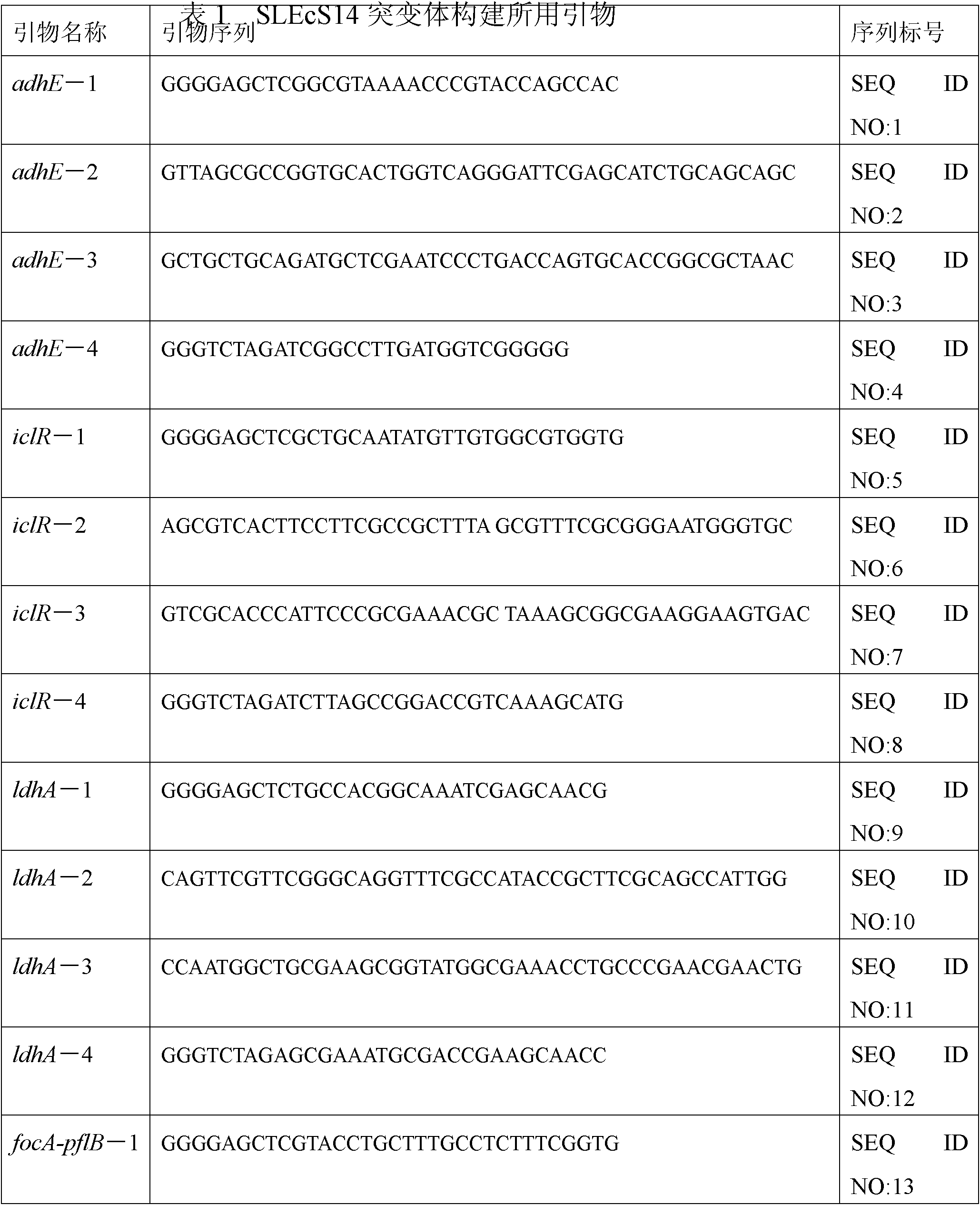
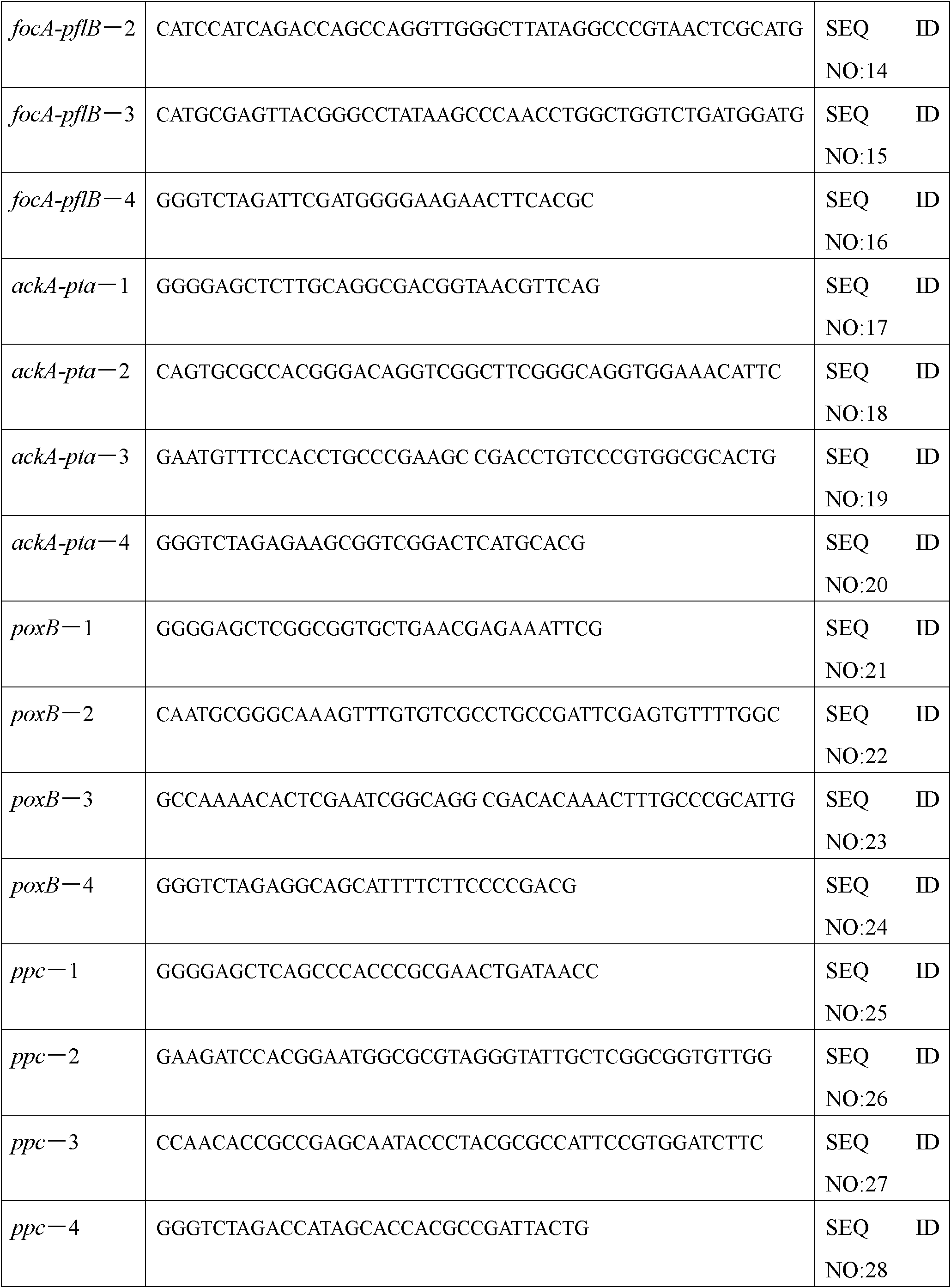
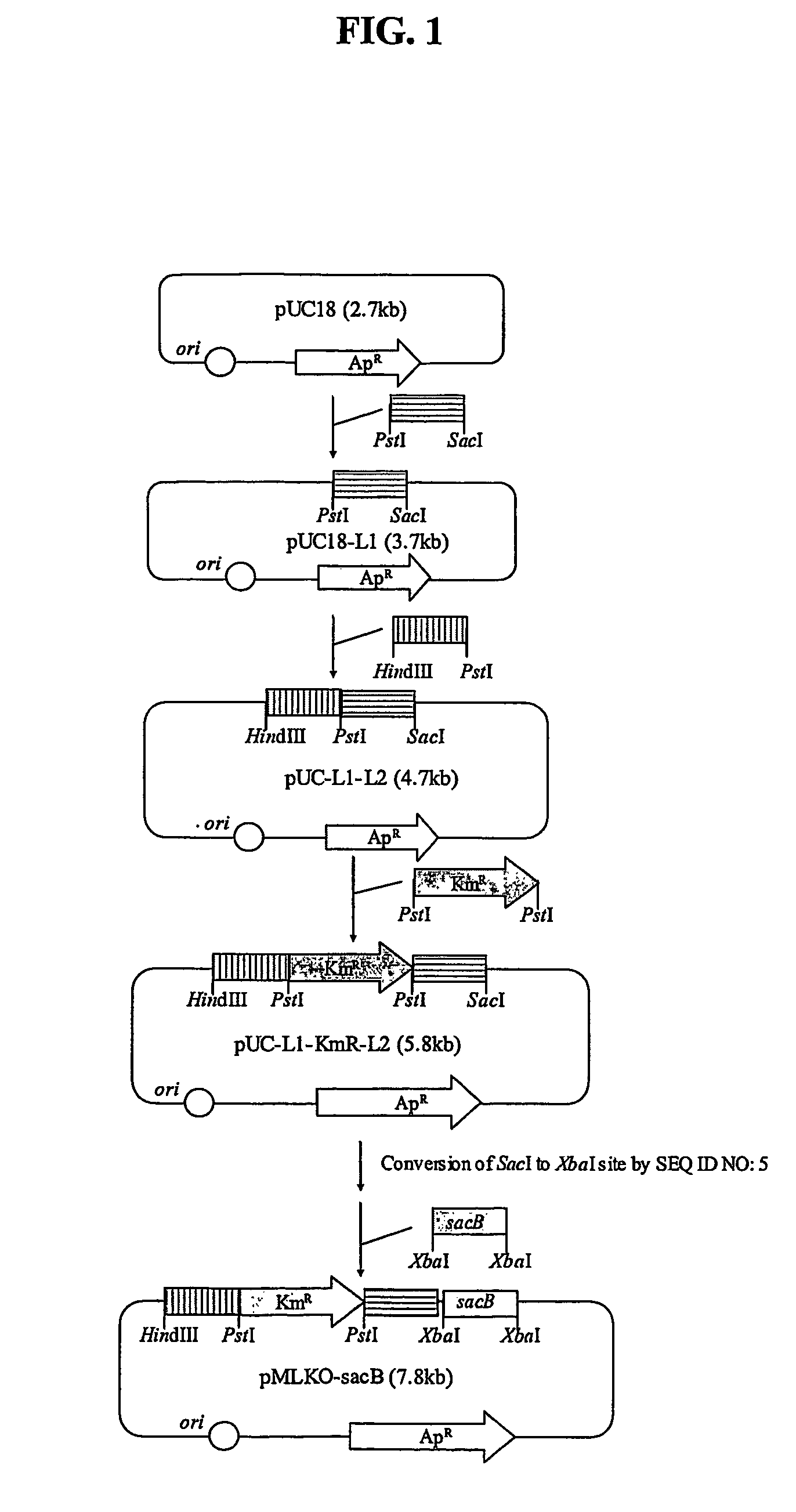
The invention relates to a bacterial strain for the high yield of succinic acid and application thereof. Escherichia coli is modified in order to remove or decrease the activities of adhE, ackA, pta, focA, pflB, iclR, ldhA, poxB and pepc genes; and the gene knockout bacterial strain then receives ultraviolet mutagenesis, and is screened by culture medium with glucose as the only carbon source, so that an engineering bacterial strain which can produce the succinic acid on basal medium and with the glucose as the only carbon source by anaerobic fermentation can be obtained. The invention adopts the succinic acid-producing escherichia coli as the starting strain, and by means of a rational design, the inveniton obtains the engineering bacterial strain with the glucose as the only carbon source theoretically, which carries out anaerobic fermentation in the basal medium and only produces the succinic acid and no other byproducts. Then by means of irrational metabolic evolution integrating ultraviolet mutagenesis with growth screening, a mutant strain for producing the succinic acid is obtained. The bacterial strain disclosed by the invention nearly produces no other heteroacids in the fermentation of the succinic acid, so the production cost of the succinic acid can be remarkably reduced.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
Rumen bacteria variants and process for preparing succinic acid employing the same
ActiveUS7470530B2High yieldSugar derivativesBacteriaHigh concentrationPhosphoenolpyruvate carboxylase
Provided are novel rumen bacterial mutants resulted from the disruption of a lactate dehydrogenase gene (ldhA) and a pyruvate formate-lyase gene (pfl) from rumen bacteria; a novel bacterial mutant (Mannheimia sp. LPK7) having disruptions of a ldhA, a pfl,a phosphotransacetylase gene (pta), and a acetate kinase gene (ackA); a novel bacterial mutant (Mannheimia sp. LPK4) having disruptions of a ldhA, a pfl, and a phosphoenolpyruvate carboxylase gene (ppc) involved in the immobilization of CO2 in a metabolic pathway of producing succinic acid; and a method for producing succinic acid, characterized by culture of the above mutants in anaerobic conditions. The bacterial mutants have the property of producing succinic acid at high concentration while producing little or no organic acids, as compared to the prior wild-type strains of producing various organic acids. Thus, the bacterial mutants are useful as strains for the industrial production of succinic acid.
Owner:KOREA ADVANCED INST OF SCI & TECH
Biocatalyst for production of d-lactic acid (as amended)
ActiveUS20070065930A1Increase productivityHigh selectivityBacteriaFermentationEscherichia coliD-lactate dehydrogenase
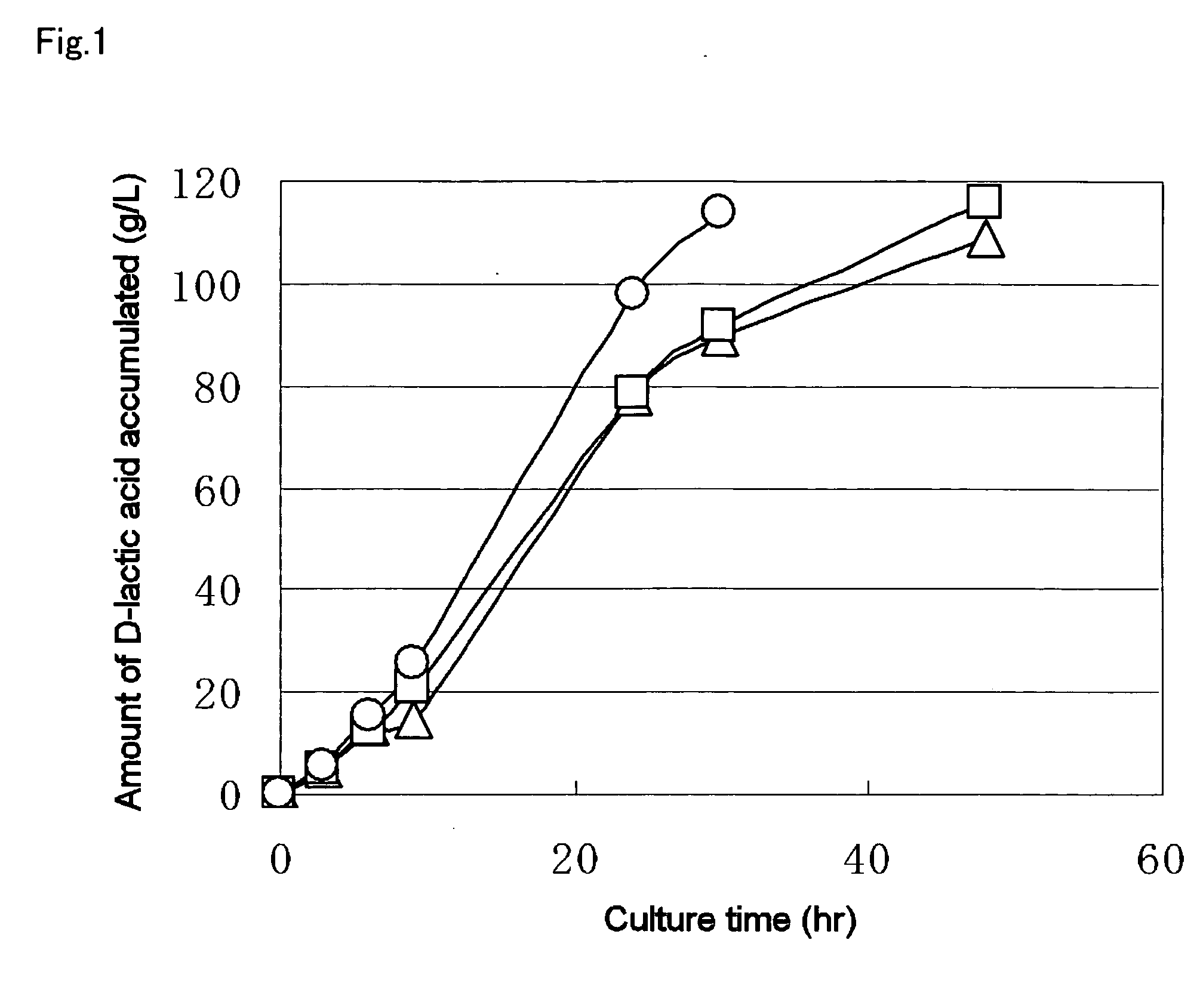
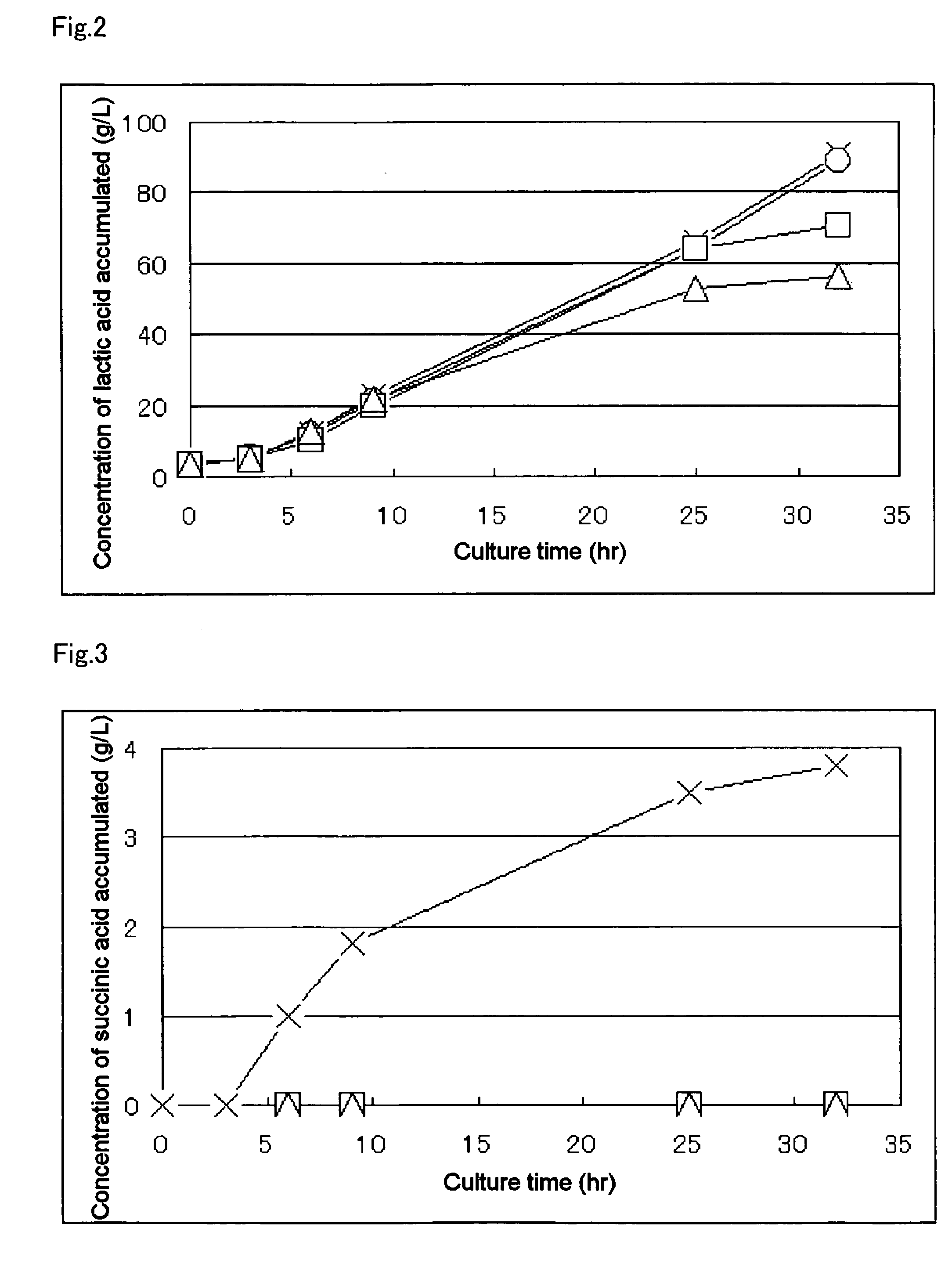
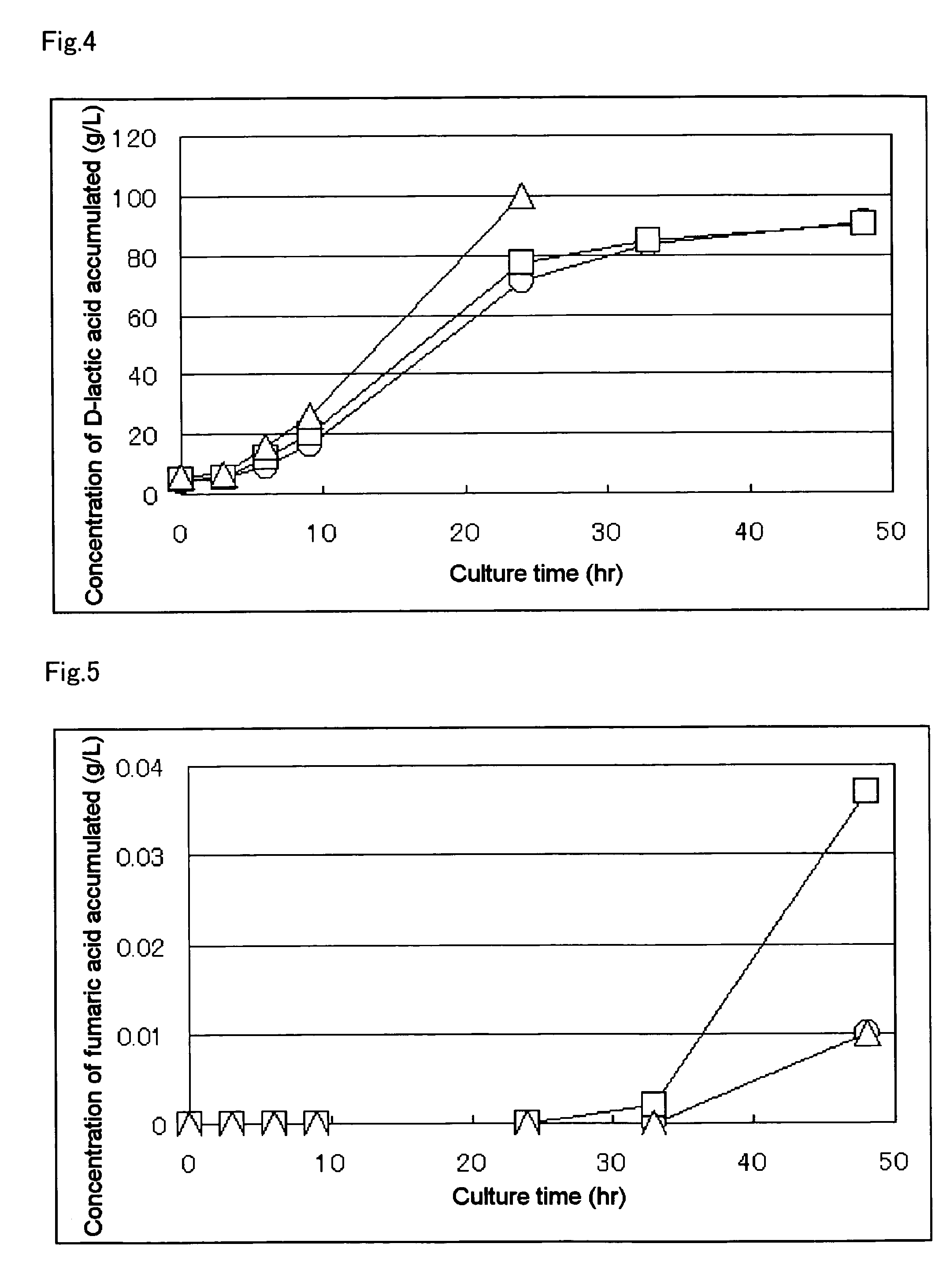
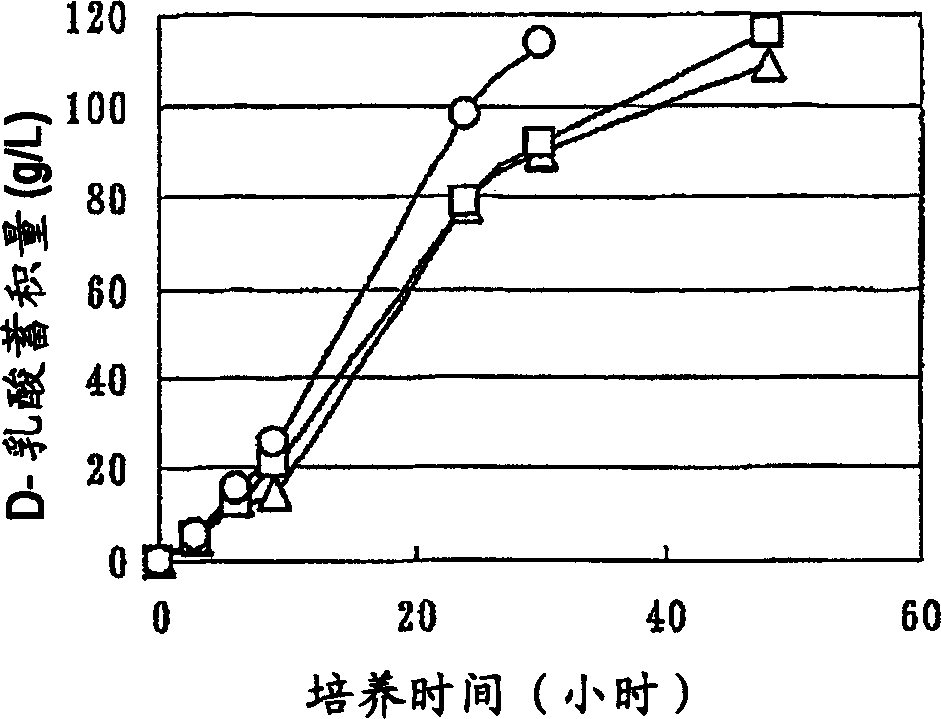
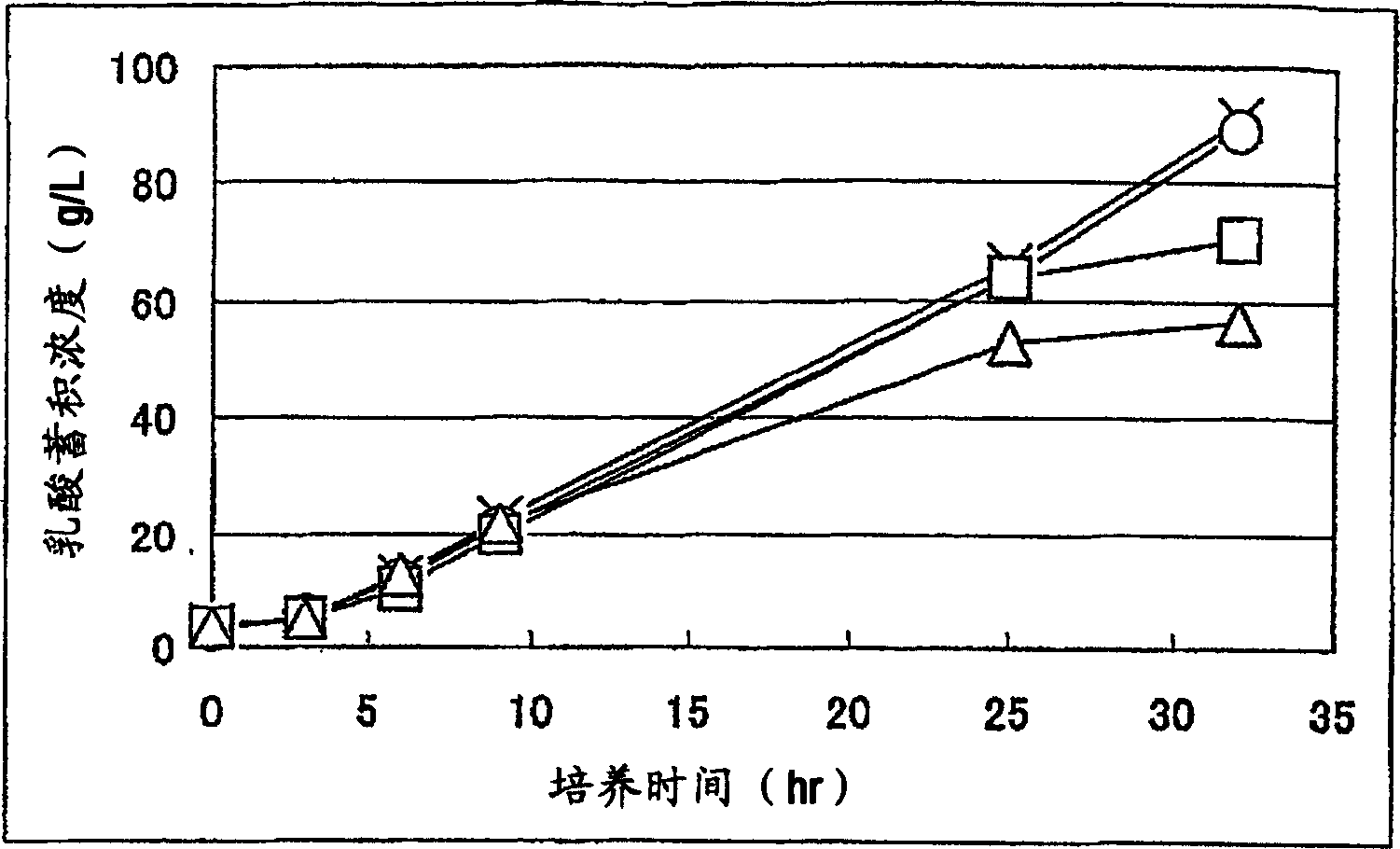
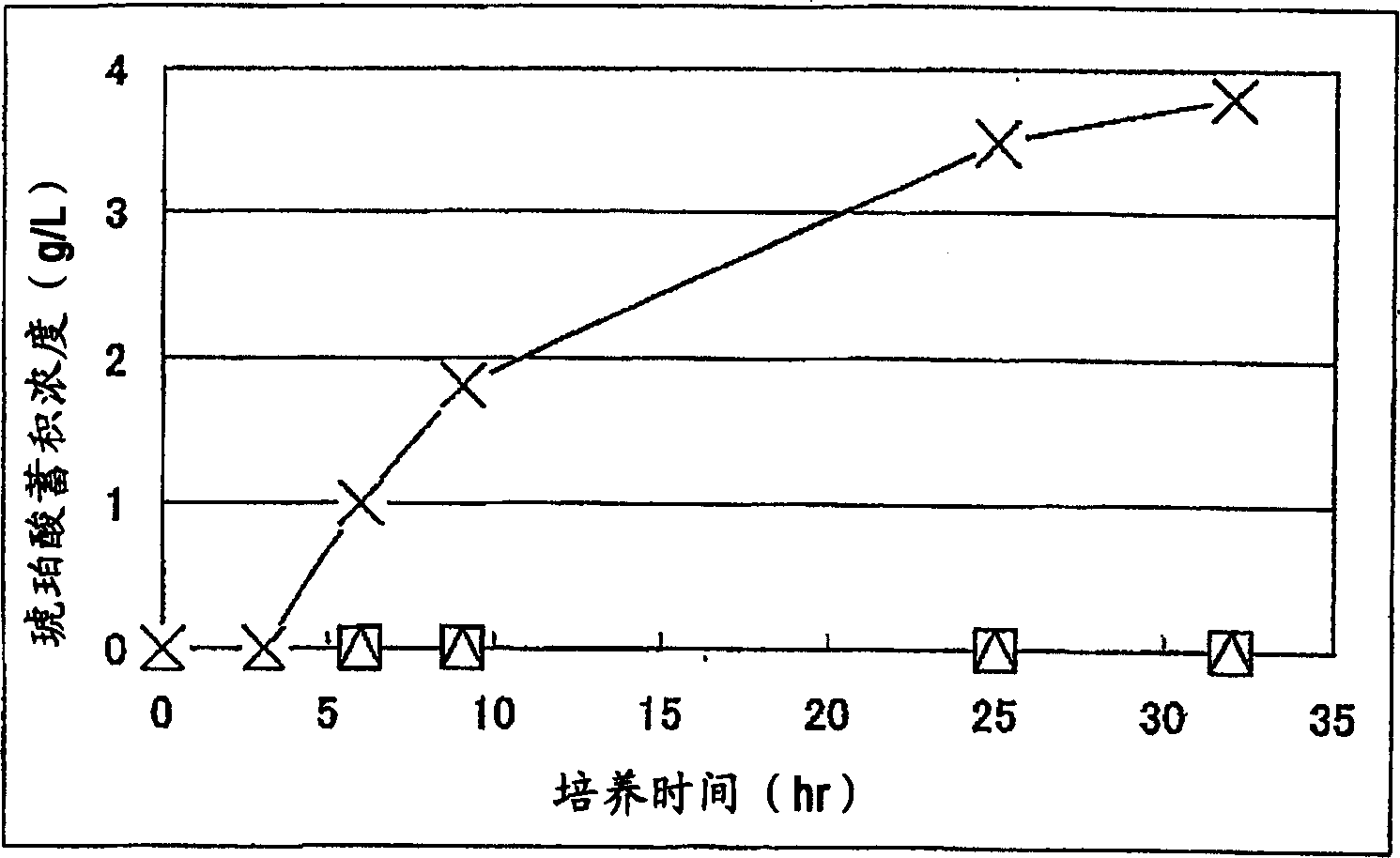
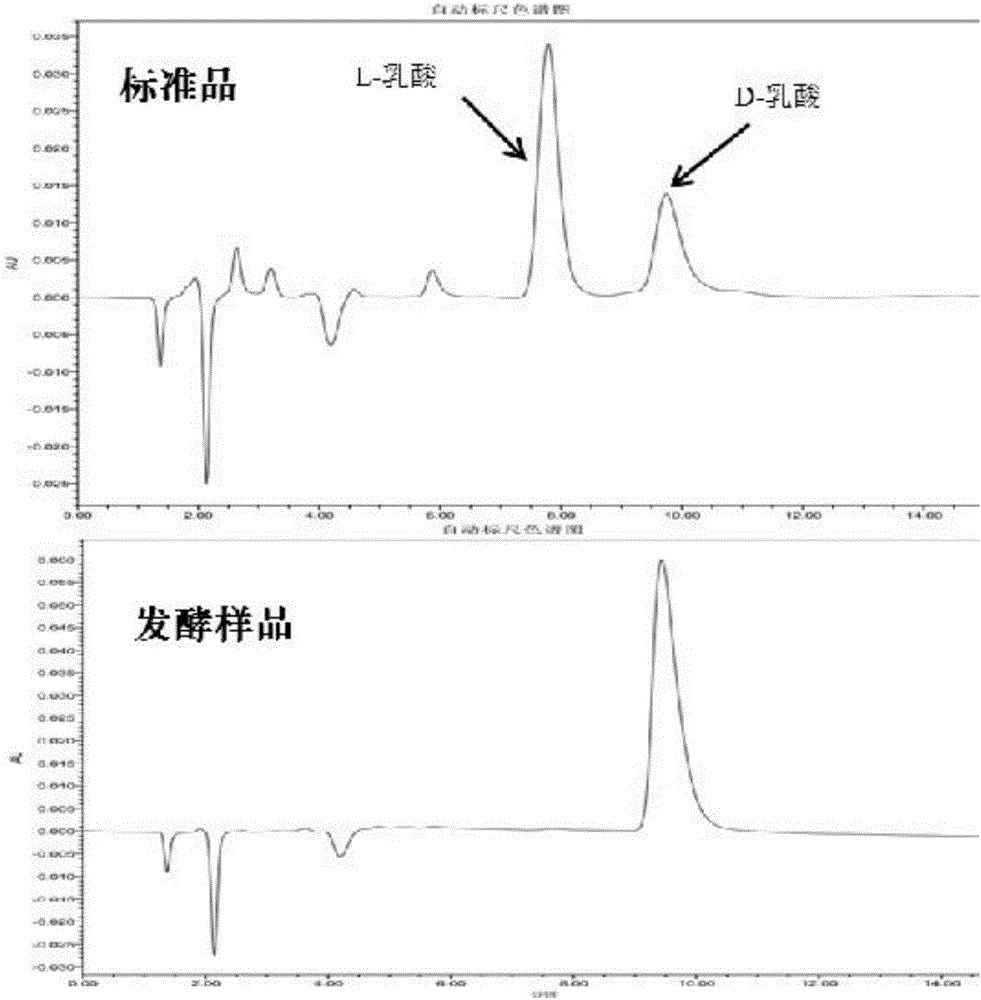
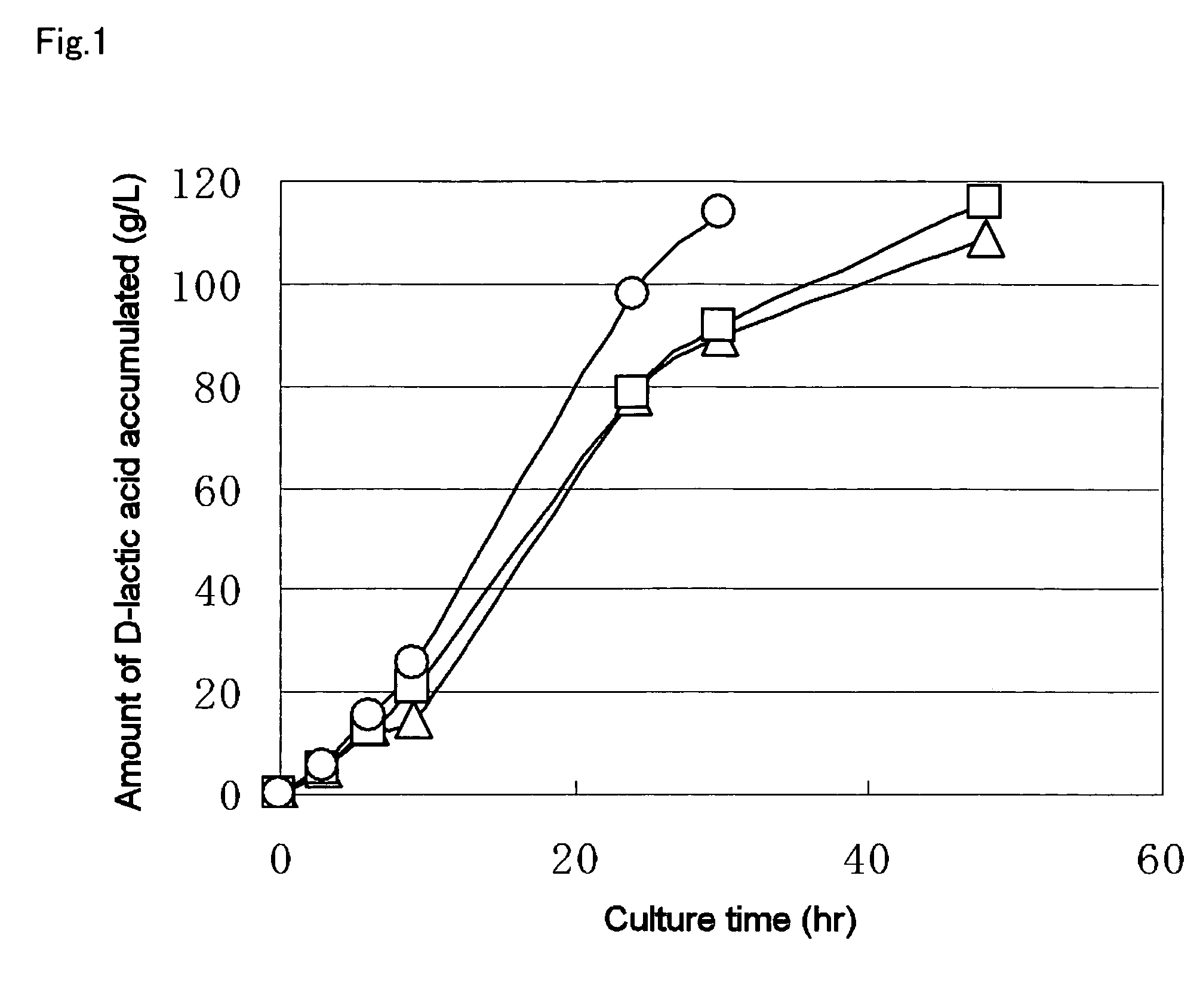
A method for producing D-lactic acid in high yield, and to provide a method for producing D-lactic acid with high selectivity, in which optical purity is high and a by-product organic acid is small. In one aspect, a microorganism, wherein activity of pyruvate formate-lyase (pfl) is inactivated or decreased, and further activity of Escherichia coli-derived NADH-dependent D-lactate dehydrogenase (ldhA) is enhanced, is cultured to efficiently produce D-lactic acid. With regard to a method for enhancing ldhA activity, by linking, on a genome, a gene encoding ldhA with a promoter of a gene which controls expression of a protein involved in a glycolytic pathway, a nucleic acid biosynthesis pathway or an amino acid biosynthesis pathway, suitable results are obtained compared to the method for enhancing expression of the gene using an expression vector. A microorganism in which a dld gene is substantially inactivated or decreased is cultured to produce high quality D-lactic acid with reduced concentration of pyruvic acid.
Owner:MITSUI CHEM INC
Biocatalyst for producing d-lactic acid
ActiveCN1856577AImprove productivityHigh purityBacteriaRecombinant DNA-technologyD-lactic acid dehydrogenase activityEscherichia coli
It is intended to provide a process for highly producing D-lactic acid. It is also intended to provide a process for highly selectively producing D-lactic acid having a high optical purity with the formation of little organic acids as by-products. D-Lactic acid is produced by culturing a microorganism having inactivated or lowered pyruvate formate-lyase activity and elevated Escherichia coli-origin NADH-dependent D-lactic acid dehydrogenase (ldhA) activity, a microorganism having inactivated or lowered FAD-dependent D-lactic acid dehydrogenase activity, or a microorganism as described above having a TCA cycle, inactivated or lowered malic acid dehydrogenase activity and inactivated or lowered aspartic acid ammonia-lyase activity. The ldhA activity is elevated by ligating a gene encoding ldhA to a promoter of a gene controlling the expression of a protein, which participates in a glycolysis system, a nucleic acid biosynthesis system or an amino acid biosynthesis system, on genome.
Owner:MITSUI CHEM INC
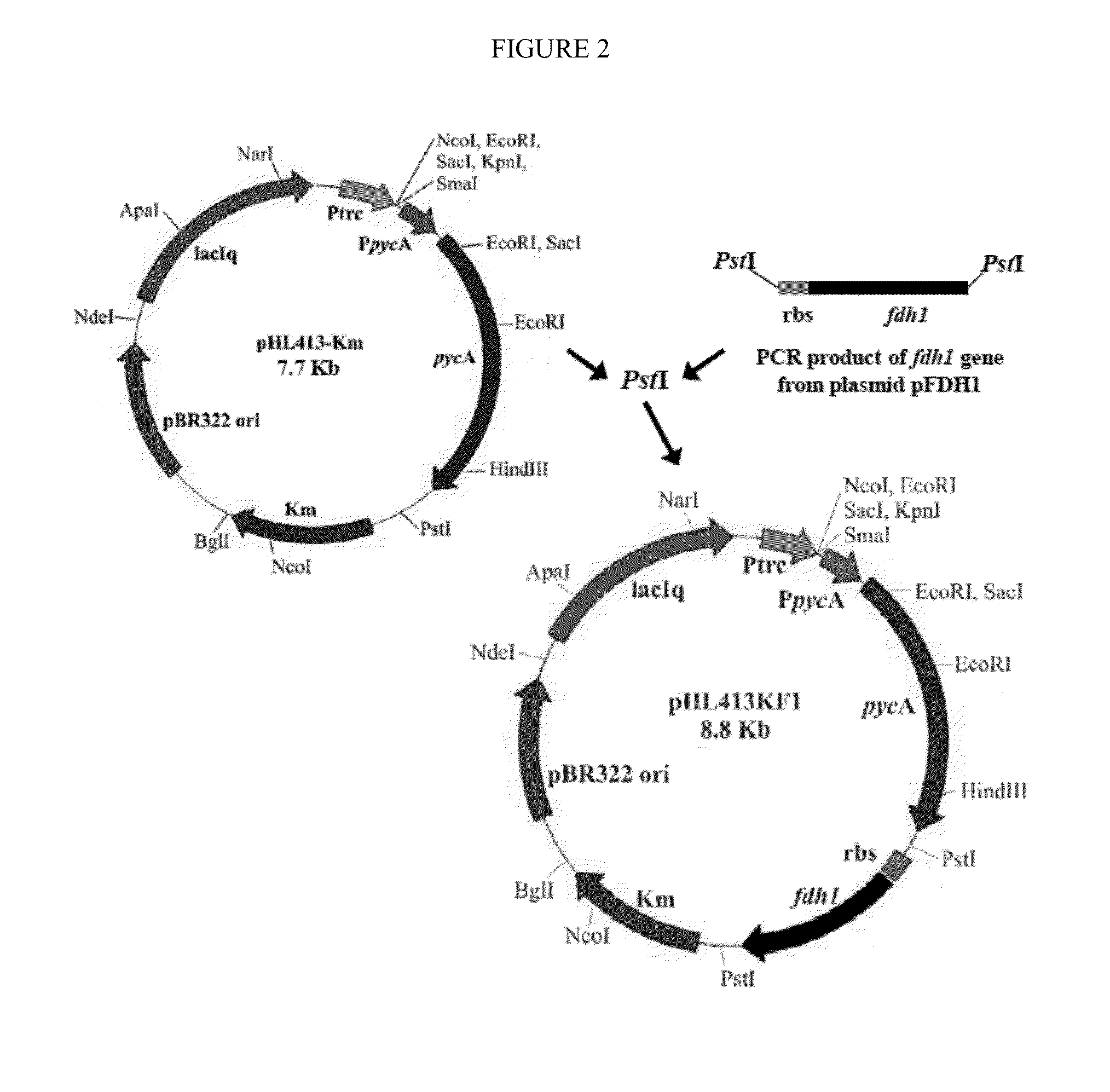
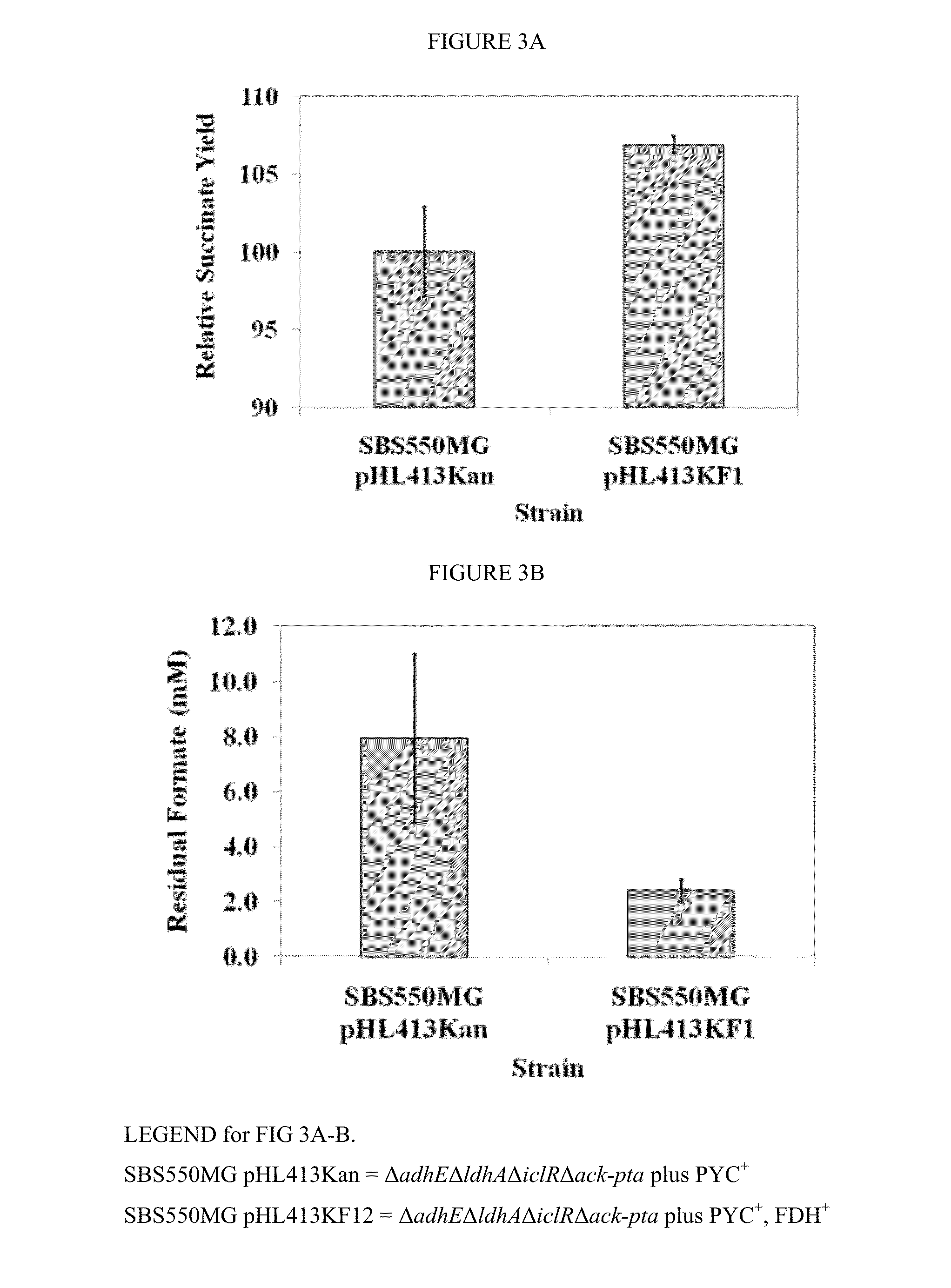
Increasing bacterial succinate productivity
ActiveUS8795991B2Increase productionReduces the byproduct formateBacteriaOxidoreductasesThe Krebs CycleMicrobiology
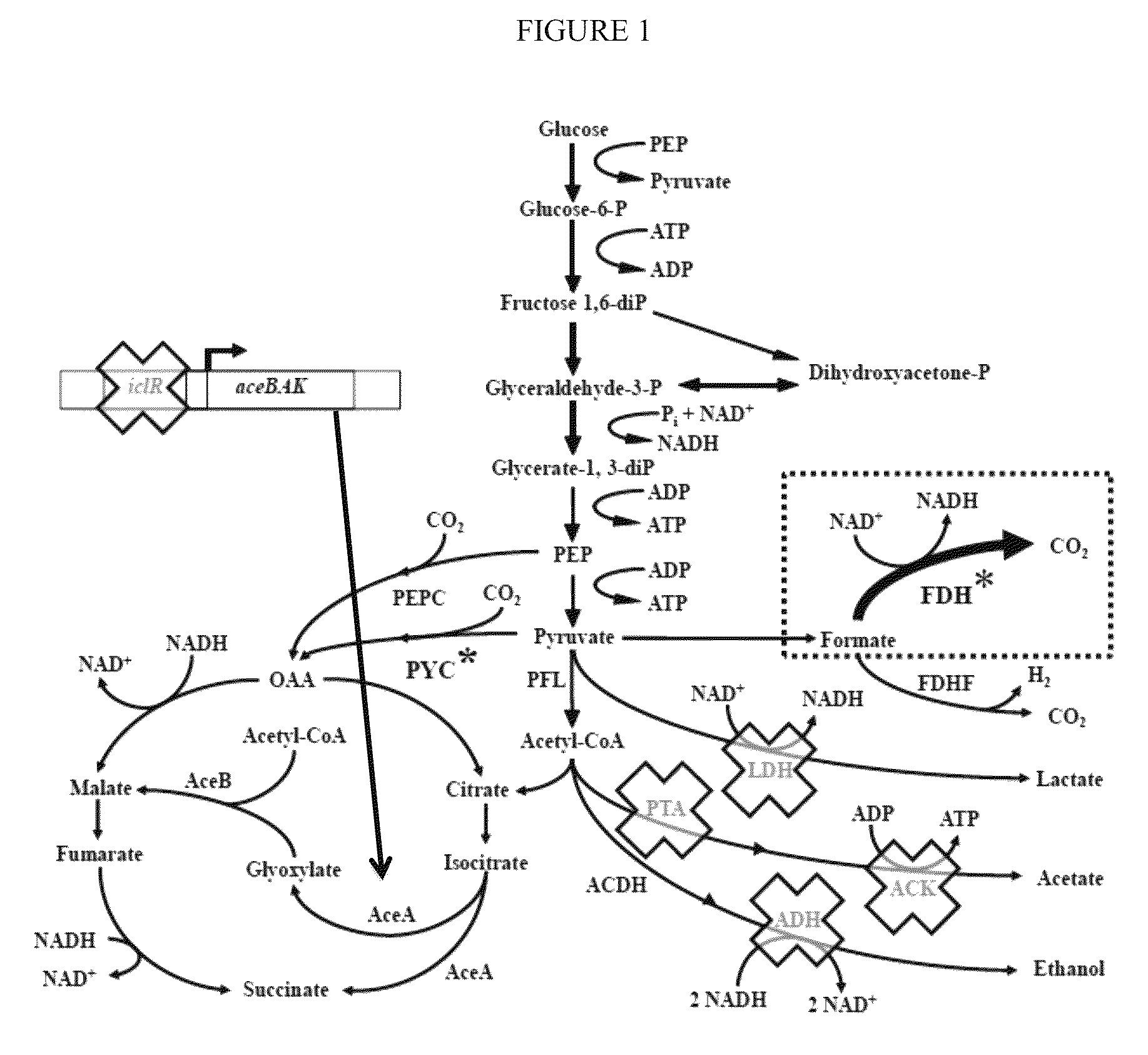
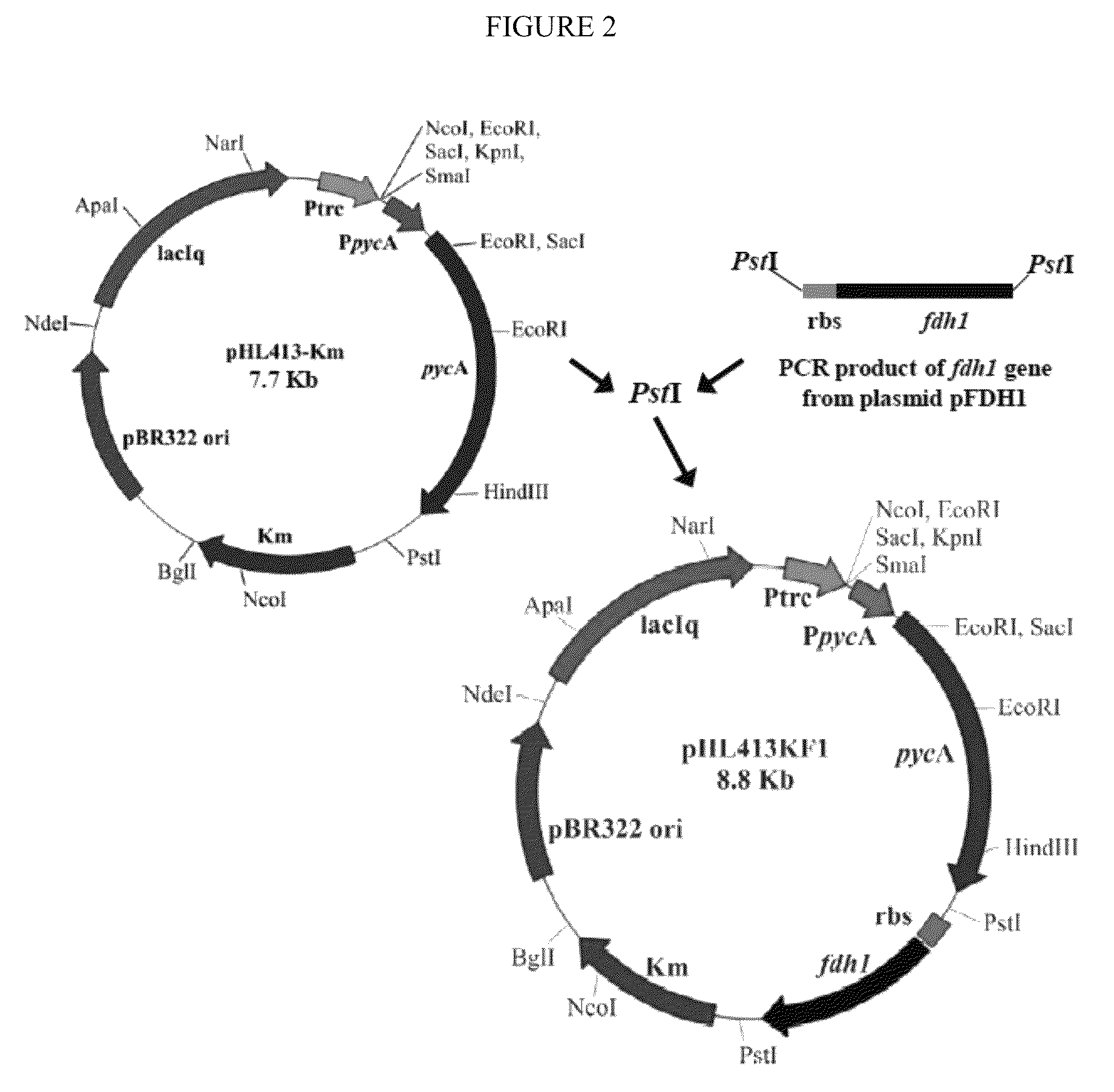
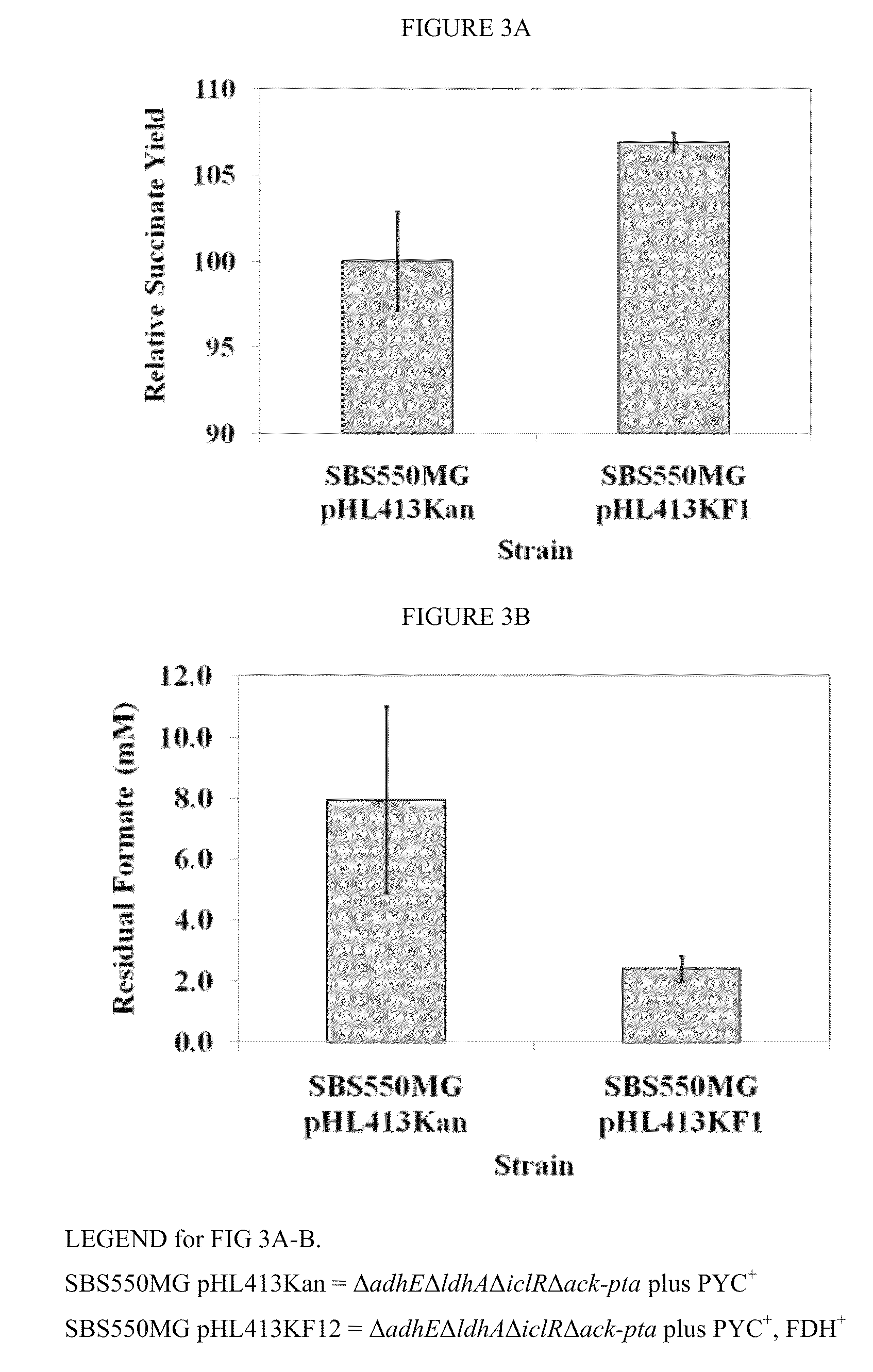
Improved bacteria for making succinate and other 4 carbon dicarboxylates from the Krebs cycle have modifications to reduce acetate, lactate, EtOH and formate, as well as turn on the glyoxylate shunt, produce more NADH and overexpress In one embodiment, the bacteria are ΔadhEΔldhAΔiclRΔack-pta plus PYC+ and NAD+-dependant FDH+.
Owner:RICE UNIV
Novel rumen bacteria variants and process for preparing succinic acid employing the same
The present invention relates to novel rumen bacterial mutants resulted from the disruption of a lactate dehydrogenase gene (ldhA) and a pyruvate formate-lyase gene (pfl) (which are involved in the production of lactic acid, formic acid and acetic acid) from rumen bacteria; a novel bacterial mutant (Mannheimia sp. LPK7) having disruptions of a lactate dehydrogenase gene (ldhA), a pyruvate formate-lyase gene (pfl), a phosphotransacetylase gene (pta), and a acetate kinase gene (ackA); a novel bacterial mutant (Mannheimia sp. LPK4) having disruptions of a lactate dehydrogenase gene (ldhA), a pyruvate formate-lyase gene (pfl) and a phosphoenolpyruvate carboxylase gene (ppc) involved in the immobilization of CO2 in a metabolic pathway of producing succinic acid; and a method for producing succinic acid, which is characterized by the culture of the above mutants in anaerobic conditions. The inventive bacterial mutants have the property of producing succinic acid at high concentration while producing little or no organic acids, as compared to the prior wild-type strains of producing various organic acids. Thus, the inventive bacterial mutants are useful as strains for the industrial production of succinic acid.
Owner:KOREA ADVANCED INST OF SCI & TECH
Novel engineered microorganism producing homo-succinic acid and method for preparing succinic acid using the same
ActiveUS20090203095A1High yieldIncrease productivityBacteriaOther foreign material introduction processesHigh concentrationMannheimia
The present invention relates to a mutant microorganism, which is selected from the group consisting of genus Mannheimia, genus Actinobacillus and genus Anaerobiospirillum, producing homo-succinic acid and a method for producing homo-succinic acid using the same, and more particularly to a mutant microorganism producing succinic acid at a high concentration while producing little or no other organic acids in anaerobic conditions, which is obtained by disrupting a gene encoding lactate dehydrogenase (ldhA), a gene encoding phosphotransacetylase (pta), and a gene encoding acetate kinase (ackA), without disrupting a gene encoding pyruvate formate lyase (pfl), as well as a method for producing succinic acid using the same. The inventive mutant microorganism has the property of having a high growth rate and succinic acid productivity while producing little or no organic acids, as compared to the prior strains producing succinic acid. Thus, the inventive mutant microorganism is useful to produce succinic acid for industrial use.
Owner:KOREA ADVANCED INST OF SCI & TECH
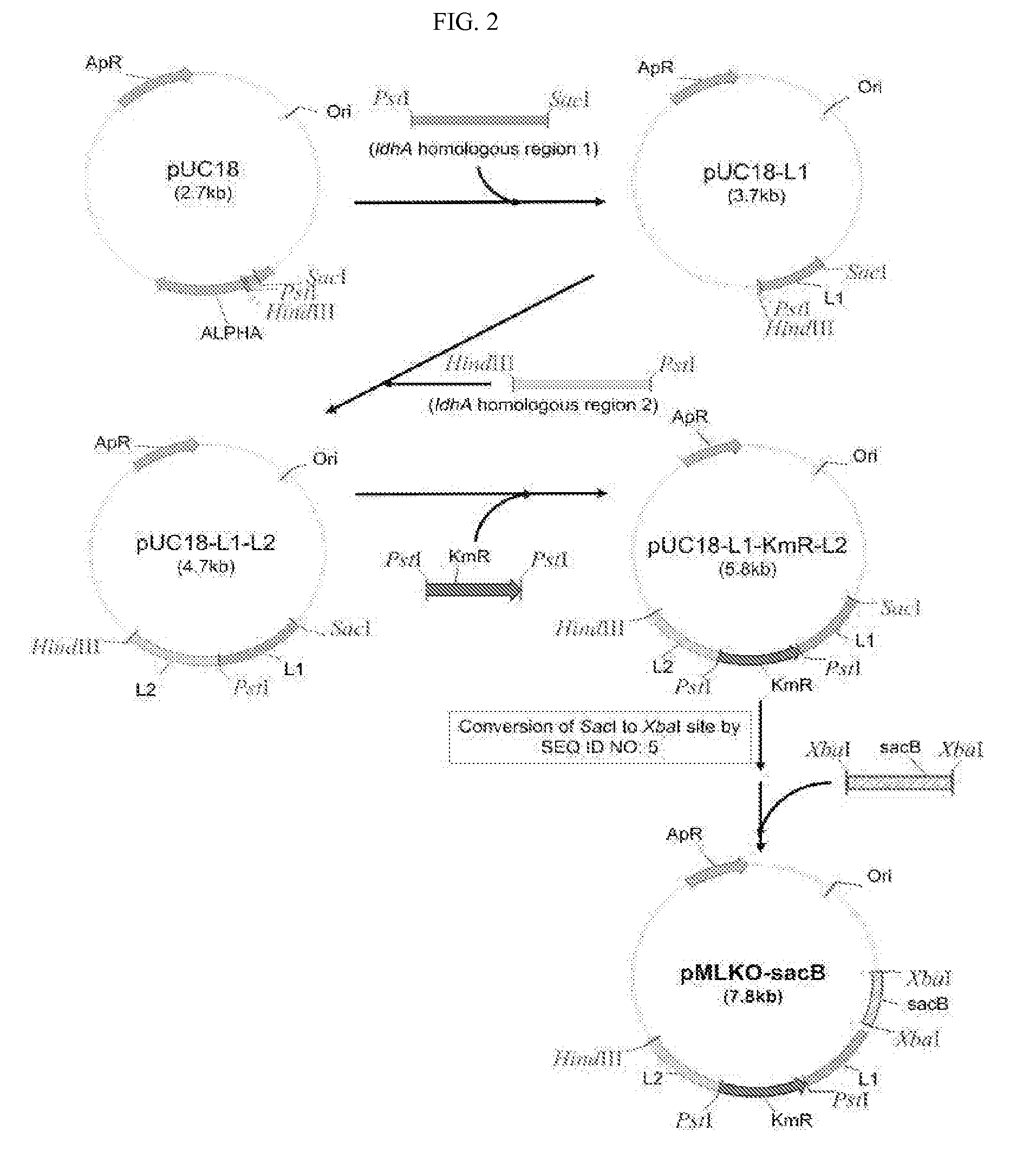
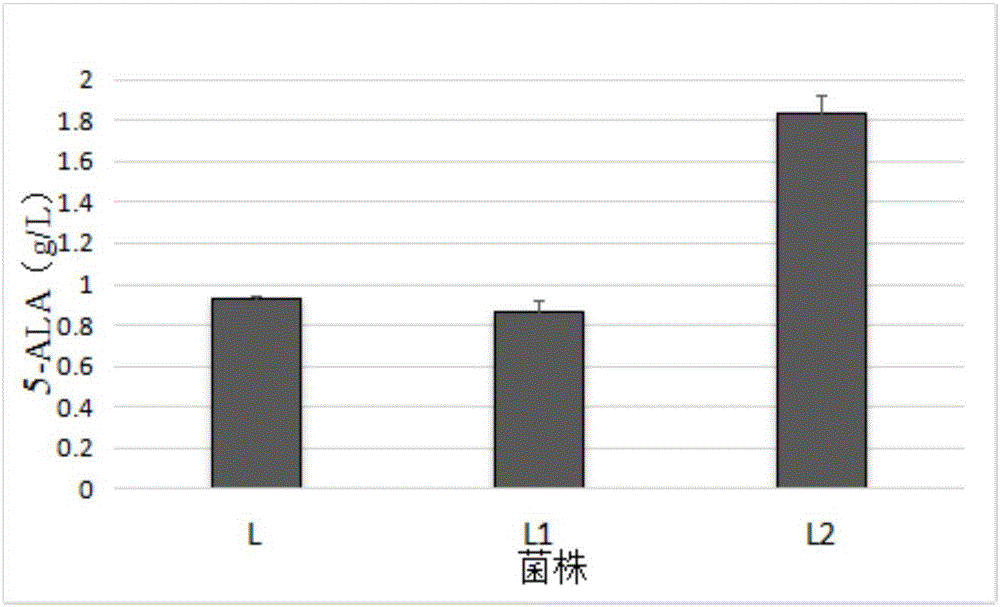
Corynebacterium glutamicum recombinant strain for producing 5-aminolevulinic acid
InactiveCN106434513AEasy to transportIncrease productionBacteriaMicroorganism based processesPhosphoenolpyruvate carboxylaseAminolevulinic acid synthase
The invention discloses a corynebacterium glutamicum recombinant strain for producing 5-aminolevulinic acid. An establishing method comprises steps as follows: (1) a lactic dehydrogenase encoding gene ldhA and acetic acid producing genes pta-ackA, pqo and cat are knocked out from corynebacterium glutamicum ATCC 1303, and a strain CG4 is obtained; an sod promoter is inserted in front of a phosphoenolpyruvate carboxylase encoding gene in the CG4, and a strain CG5 is obtained; a phosphoenolpyruvate carboxykinase encoding gene pck is knocked out from the CG5, and a strain CG6 is obtained; (2) plasmids of an overexpressed 5-aminolevulinic acid synthase gene are transferred into the CG6, and a recombinant strain L is obtained; (3) 5-aminolevulinic acid transport protein plasmids are transferred into the L. The recombinant strain can promote 5-aminolevulinic acid to be transported outside corynebacterium glutamicum cells, and the yield of 5-aminolevulinic acid is 112.3% higher than that of a contrast strain.
Owner:TIANJIN UNIV
Genetically engineered bacterium with high-yield electroactivity and environmental stress tolerance
ActiveCN106520653AImprove electricity production activityHigh electroactivityBacteriaMicroorganism based processesBiotechnologySynechococcus
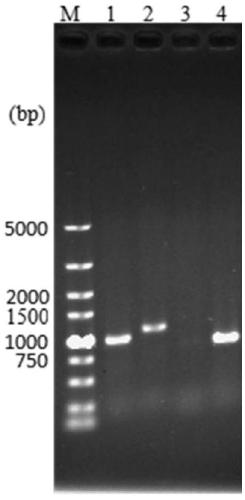
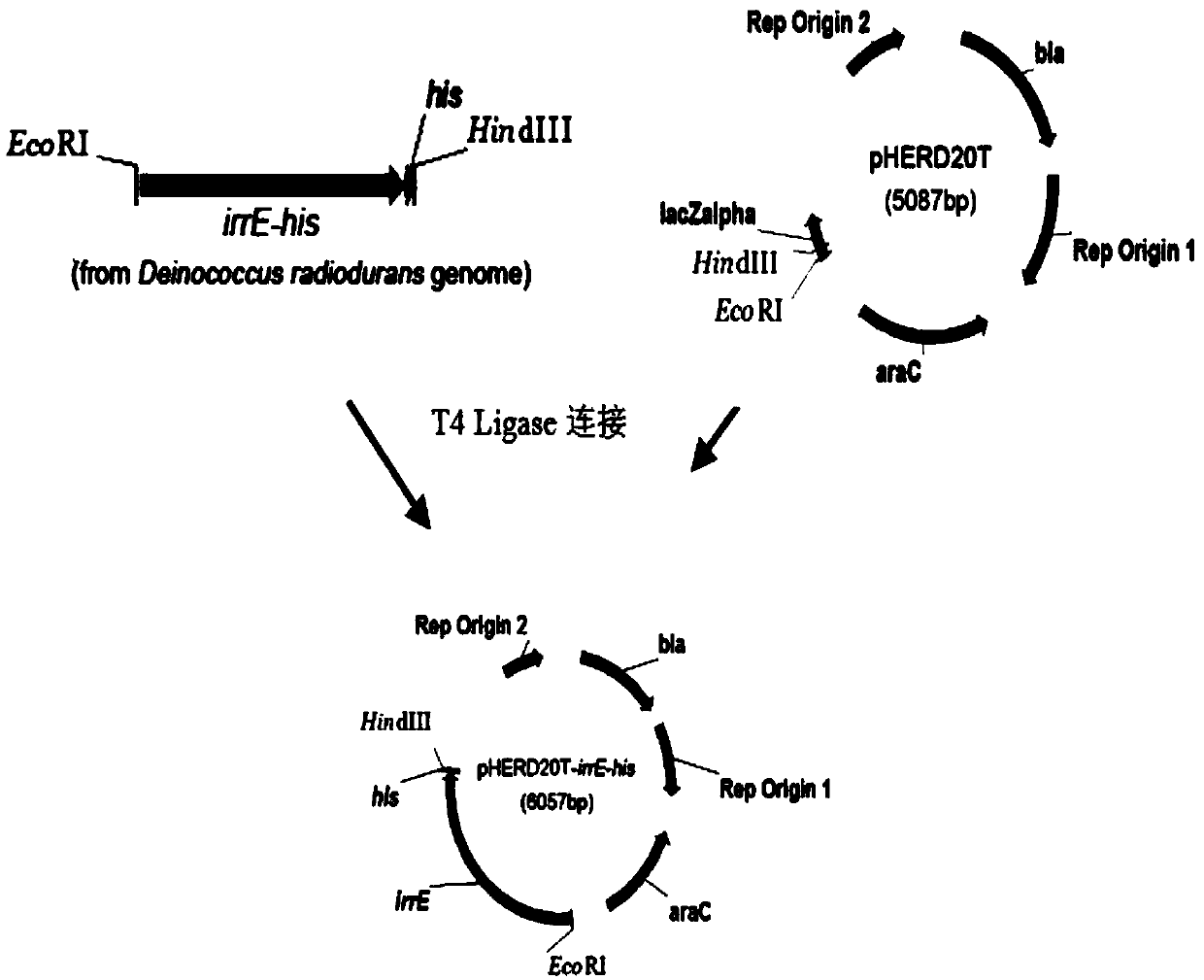
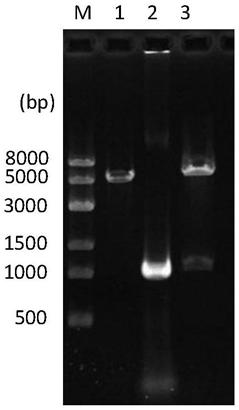
The invention relates to a genetically engineered bacterium with high-yield electroactivity and environmental stress tolerance. The lactic dehydrogenase gene ldhA in a pseudomonas aeruginosa PAO1 genome is knocked out, and the PAOl knockout strain is obtained; then a global regulatory factor IrrE of the deinococcus radiodurans is guided into the ldhA-, and the genetic engineering strain ldhA--irrE is obtained. After the thallus treated through a chemical agent is inoculated to a microbial fuel cell, the generated voltage and power density are improved by 46.76%-53.33% than those of the wild type strain, the time for stabilizing a system is shortened by 28.57%, the internal resistance of the system is reduced by 12.66%, the voltage under polyethylene glycol treatment is 543, the power density is 207, the stabilizing time is 130 h, the internal resistance reaches 420.32, and the survival rate of the engineered strain under the hunger condition, the high-acid-base condition and high-salt condition are improved by one time than those of the wild type strain.
Owner:TIANJIN UNIVERSITY OF SCIENCE AND TECHNOLOGY
Genetic engineering bacteria for producing L-lactic acid from acetic acid, construction method thereof and application thereof
ActiveCN109321590AIncrease productionReduce usageBacteriaMicroorganism based processesBiotechnologyAcetic acid
The invention discloses genetic engineering bacteria for producing L-lactic acid from acetic acid, a construction method thereof and application thereof. The genetic engineering bacteria for producingL-lactic acid from acetic acid are obtained by modifying the recipient bacteria as follows: knocking out the poxB gene, pflB gene, aceEF gene, ldhA gene and lldD gene of the recipient bacteria; and increasing the content of protein encoded by the acs gene, pta gene, ackA gene, aceA gene, aceK gene, aceB gene, maeA gene, maeB gene, and ldh2 gene in the recipient bacteria. According to the engineering bacteria obtained by the invention, the desired level of the yield of L-lactic acid obtained by using acetic acid in shake flask culture is achieved, the use of carbon sources such as glucose is avoided, the reduction of the raw material cost of L-lactic acid fermentation is facilitated, and good application prospects are realized.
Owner:BEIJING UNIV OF CHEM TECH
Recombinant Escherichia coli for synthesizing S-1,2-propanediol from L-lactic acid and construction method thereof
ActiveCN104109651AIncrease productionImprove conversion rateBacteriaMicroorganism based processesEscherichia coliSodium lactate
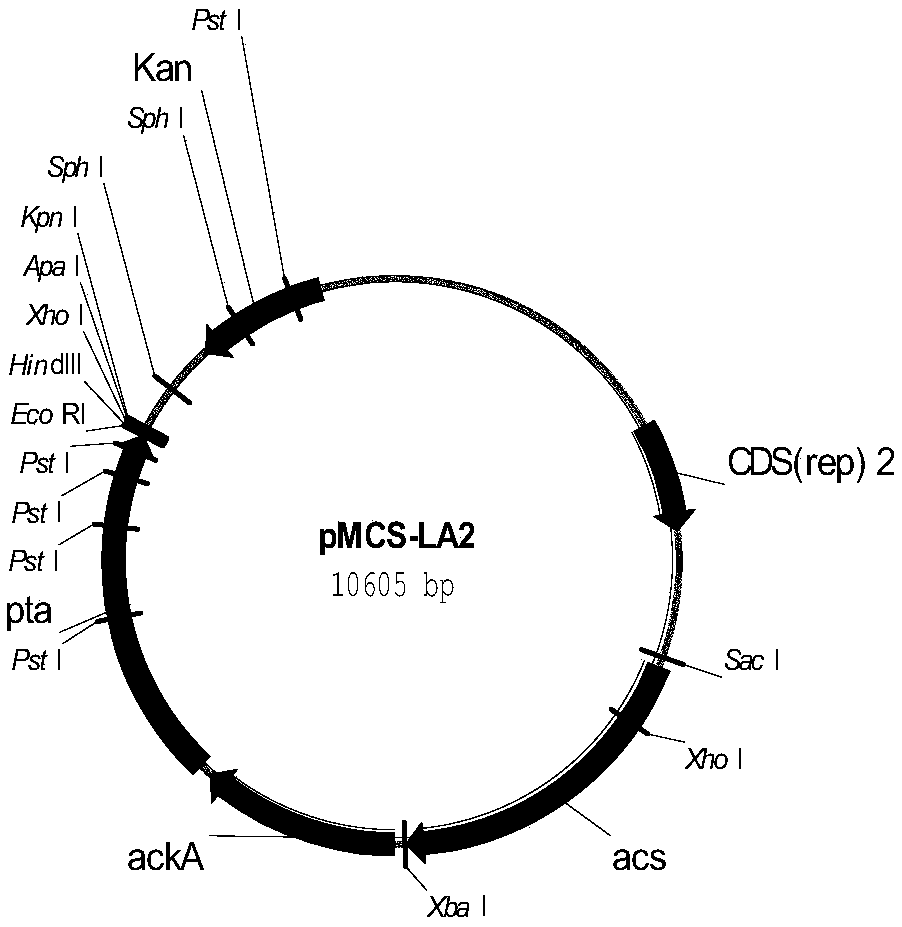
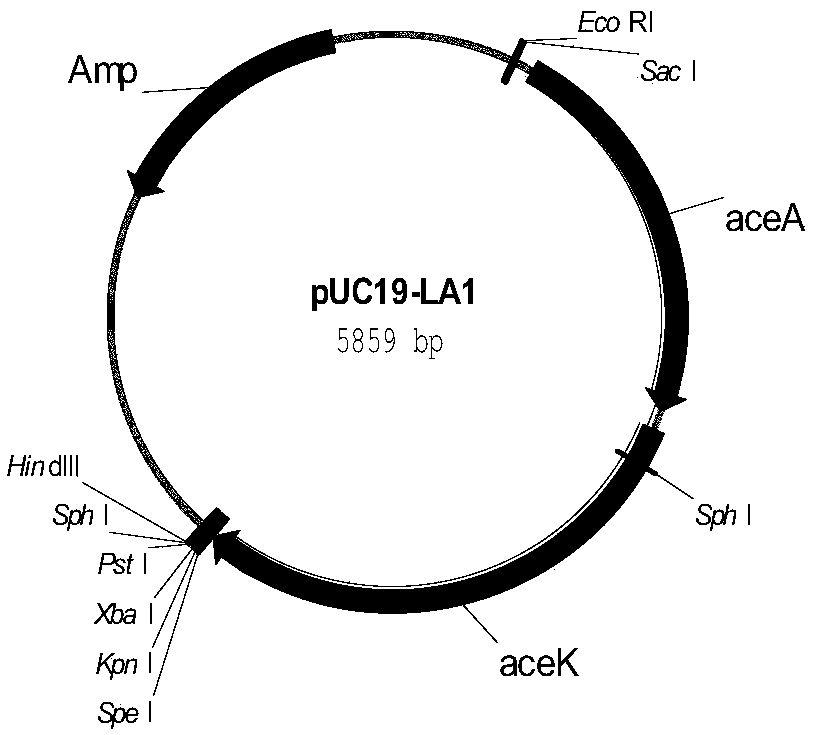
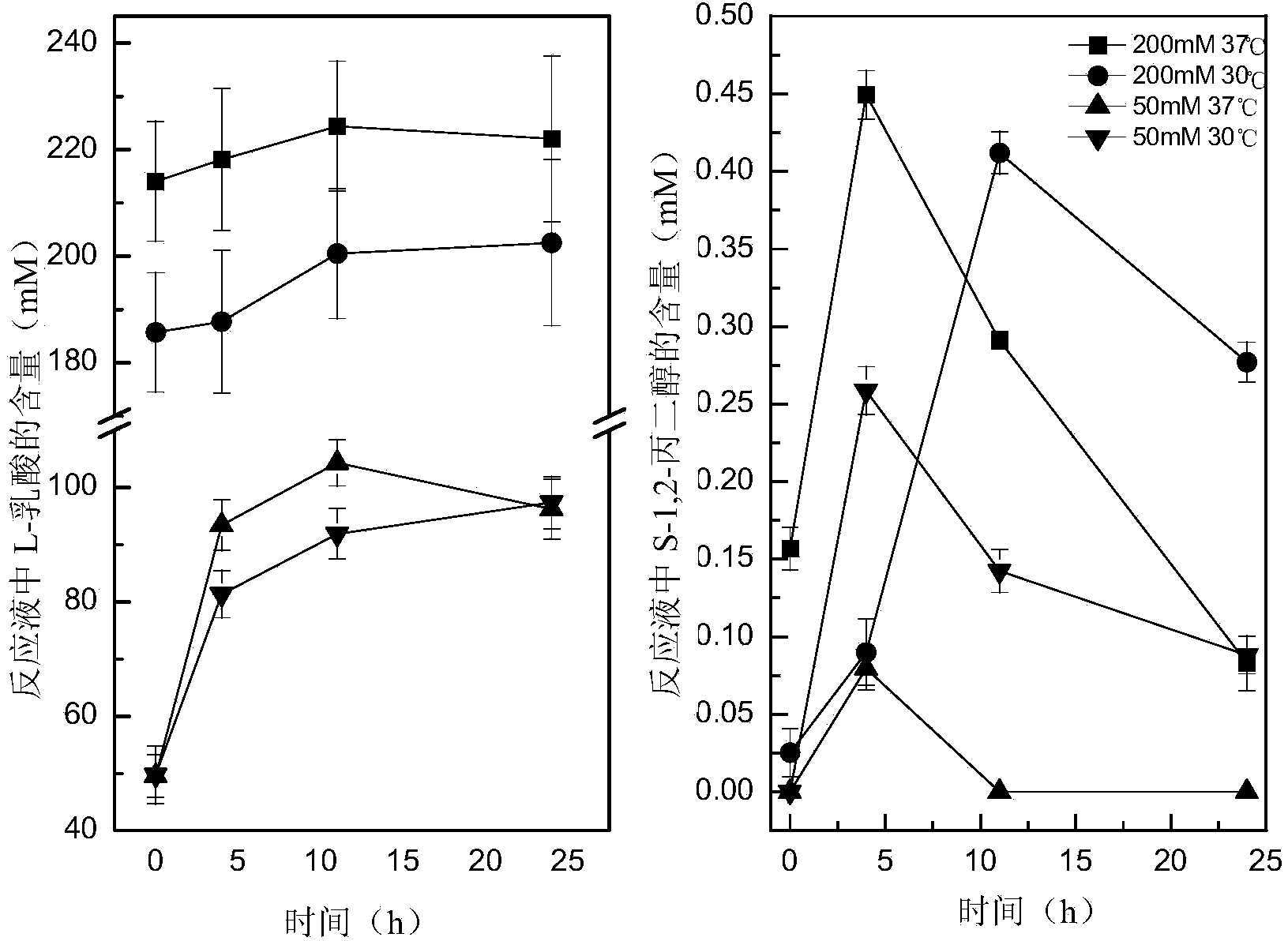
The invention discloses a recombinant Escherichia coli for synthesizing S-1,2-propanediol from L-lactic acid and a construction method thereof. The method comprises the following steps: 1) respectively substituting lldD gene, adheE gene and ackA-pta gene in Escherichia coli BW25113-DELTApoxB with coding gene of 3-hydroxypropionate dehydrogenase, coding gene of coenzyme A-dependent succinic semialdehyde dehydrogenase and coding gene of propionyl coenzyme A transferase to obtain recombinant bacterium BWPDO1; 2) knocking out ldhA gene and dld gene of the BWPDO1 to obtain recombinant bacterium BWPDO2; and 3) transferring coding gene of pyruvate decarboxylase and coding gene of NAD-dependent acetaldehyde coenzyme A dehydrogenase into the BWPDO2 to obtain recombinant bacterium BWPDO3 which is the recombinant Escherichia coli. The experiment proves that the recombinant Escherichia coli can generate S-1,2-propanediol by converting L-lactic acid; and by using 200 mM of sodium lactate as the substrate, the culture is performed at 37 DEG C for 24 hours to generate 3.4 mM of S-1,2-propanediol. The recombinant Escherichia coli lays foundation for enhancing the yield and conversion rate of the biosynthetic S-1,2-propanediol.
Owner:普立思生物科技有限公司
Corynebacterium glutamicum engineering strain for producing 5-aminolevulinic acid
InactiveCN106434514AEasy to transportIncrease productionBacteriaMicroorganism based processesPhosphoenolpyruvate carboxylasePyruvate carboxylase
The invention discloses a corynebacterium glutamicum engineering strain for producing 5-aminolevulinic acid. A construction method comprises steps as follows: (1) a lactate dehydrogenase coding gene ldhA and acetic acid generation genes pta-ackA, pqo and cat are knocked out from corynebacterium glutamicum ATCC 13032, and a strain CG4 is obtained; an sod promoter is inserted in front of a phosphoenolpyruvate carboxylase coding gene ppc in the strain CG4, and a strain CG5 is obtained; a phosphoenolpyruvate carboxylase coding gene pck is knocked out from the strain CG5, and a strain CG6 is obtained; (2) plasmids over-expressing 5-aminolevulinic acid synthase genes are transferred in the CG6, and an engineering strain L is obtained; (3) transport protein plasmids over-expressing 5-aminolevulinic acid are transferred to the L. The engineering strain can promote 5-aminolevulinic acid to be transported outside the corynebacterium glutamicum, and the yield of 5-aminolevulinic acid is 100.4% higher than that of a contrast strain.
Owner:TIANJIN UNIV
Biocatalyst for production of d-lactic acid
ActiveCN101654666AImprove productivityHigh purityBacteriaMicroorganism based processesEscherichia coliSuccinic acid
The subject of the present invention is to provide a method for producing D-lactic acid in high yield, and to provide a method for producing D-lactic acid with high selectivity, in which optical purity is high and a by-product organic acid is small. A microorganism, wherein activity of pyruvate formate-lyase (pfl-) is inactivated or decreased, and further activity of Escherichia coli-derived NADH-dependent D-lactate dehydrogenase (ldhA) is enhanced, is cultured to produce a remarkable amount of D-lactic acid in a short time. With regard to a method for enhancing ldhA activity, by linking, on agenome, a gene encoding 1dhA with a promoter of a gene which controls expression of a protein involved in a glycolytic pathway, a nucleic acid biosynthesis pathway or an amino acid biosynthesis pathway, suitable results are obtained compared to the method for enhancing expression of the gene using an expression vector. In addition, a microorganism in which a did gene is substantially inactivatedor decreased is cultured to produce high quality D-lactic acid with reduced concentration of pyruvic acid. Furthermore, it is possible to suppress by-production of succinic acid and fumaric acid whilemaintaining high D-lactic acid productivity by using the above-mentioned microorganism having a TCA cycle, wherein activity of malate dehydrogenase (mdh) is inactivated or decreased, and further activity of aspartate ammonia-lyase (aspA) is inactivated or decreased.
Owner:MITSUI CHEM INC
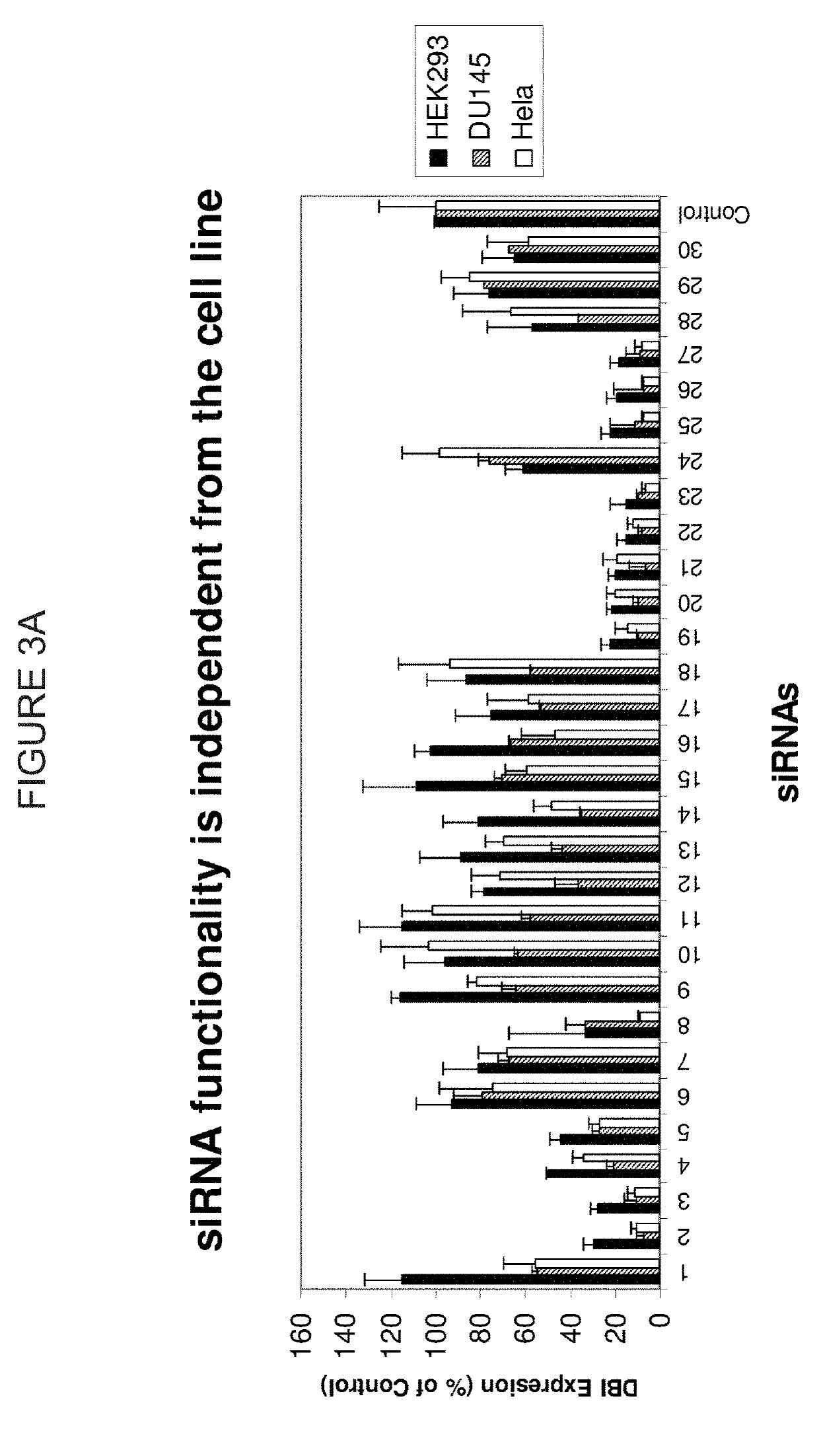
siRNA Targeting LDHA
ActiveUS20190106699A1Improve efficiencyGood curative effectScreening processDNA/RNA fragmentationNucleotideGene silencing
Efficient sequence specific gene silencing is possible through the use of siRNA technology. Be selecting particular siRNAs by rational design, one can maximize the generation of an effective gene silencing reagent, as well as methods for silencing genes. Methods compositions, and kits generated through rational design of siRNAs are disclosed, including those directed to the nucleotide sequences for LDHA.
Owner:THERMO FISHER SCIENTIFIC INC
Streptococcus zooepidemicus lactic dehydrogenase gene mutant and application thereof
ActiveCN108192899AReduce accumulationIncrease productionBacteriaOxidoreductasesStreptococcus zooepidemicusNucleotide
The invention provides a streptococcus zooepidemicus lactic dehydrogenase gene mutant and application thereof, and belongs to the technical field of microorganism gene engineering. The streptococcus zooepidemicus causes mutation of one basic group of a lactic dehydrogenase group ldhA through NTG mutagenesis, a 697-bit basic group mutates from A to G (A697G), causing that a 233-bit amino acid residue of the amino acid sequence mutates from threonine into alanine (T233A); the nucleotide sequence of the gene mutant is shown as SEQ ID No.1, so that the enzyme activity of lactic dehydrogenase can be lowered by 50 percent or more, thereby further proving that the streptococcus zooepidemicus containing the gene mutant can remarkably reduce the accumulation of lactic acid during production of hyaluronic acid through fermentation of the streptococcus zooepidemicus containing the gene mutant, the accumulation amount of lactic acid is lowered by 60 percent or more, and the yield of the hyaluronicacid is increased by 50 percent or more. The streptococcus zooepidemicus lactic dehydrogenase gene mutant has an important industrial application prospect.
Owner:TSINGHUA UNIV
Increasing bacterial succinate productivity
ActiveUS20130203137A1Increase production rateHigh yieldBacteriaOxidoreductasesMicrobiologyThe Krebs Cycle
Improved bacteria for making succinate and other 4 carbon dicarboxylates from the Krebs cycle have modifications to reduce acetate, lactate, EtOH and formate, as well as turn on the glyoxylate shunt, produce more NADH and overexpress In one embodiment, the bacteria are ΔadhEΔldhAΔiclRΔack-pta plus PYC+ and NAD+-dependant FDH+.
Owner:RICE UNIV
Lactate dehydrogenase A acetylation activator and application thereof
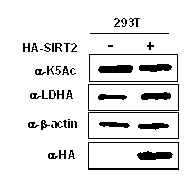
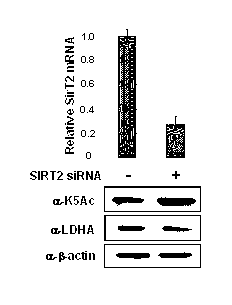
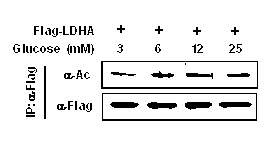
InactiveCN102805861AThe detection method is simpleEfficient detection methodPeptide/protein ingredientsAntineoplastic agentsTumor SampleAcetylation
The invention belongs to the field of molecular biology and medicine, and relates to a lactate dehydrogenase A acetylation activator and an application thereof. The lactate dehydrogenase A acetylation activator comprises an LDHA K5 acetylation activator which can be used for effectively activating acetylation of K5. The invention further provides a corresponding antibody and a method for detecting LDHA K5 acetylation. The detection method can be used for detecting the level of LDHA K5 acetylation in a tumor sample, is simple, practicable, rapid and effective, and is low in cost; and the LDHA K5 acetylation is detected by using the method, and inhibition is performed specific to the level of LDHA K5 acetylation, so that a novel simple and convenient way is provided for tumor metabolism diagnosis and treatment.
Owner:FUDAN UNIV
Genetic engineering bacteria for producing D-lactic acid and constructon method and application thereof
InactiveCN101993850BHigh optical purityIncrease production capacityBacteriaMicroorganism based processesL-Lactate dehydrogenaseMicroorganism
The invention relates to genetic engineering bacteria for producing D-lactic acid and a construction method and application thereof. The strain of C.glutamicum Res 167 delta ldh deleted in L-lactate dehydrogenase gene (ldh) is used as a starting strain to over express exogenous D-lactate dehydrogenase gene so as to obtain C.glutamicum Res 167 delta ldh / ldhA. The genetic engineering bacteria are preserved in China General Microbiological Culture Collection Center with the preserving registration number of CGMCC No.4041. By utilizing the genetic engineering means, the expression of the exogenous D-lactate dehydrogenase gene is realized in the corynebacterium glutamicum deleted in L-lactic acid metabolic pathway, and the genetic engineering bacteria producing high optical purity D-lactic acid are successfully constructed. When the engineering bacteria are used for producing lactic acid through fermentation, the yield of the D-lactic acid is over 40g / L, and the purity is over 99 percent. The invention has important significance for the industrial production of the D-lactic acid, and has wide application prospect.
Owner:FUSHUN RES INST OF PETROLEUM & PETROCHEMICALS SINOPEC CORP
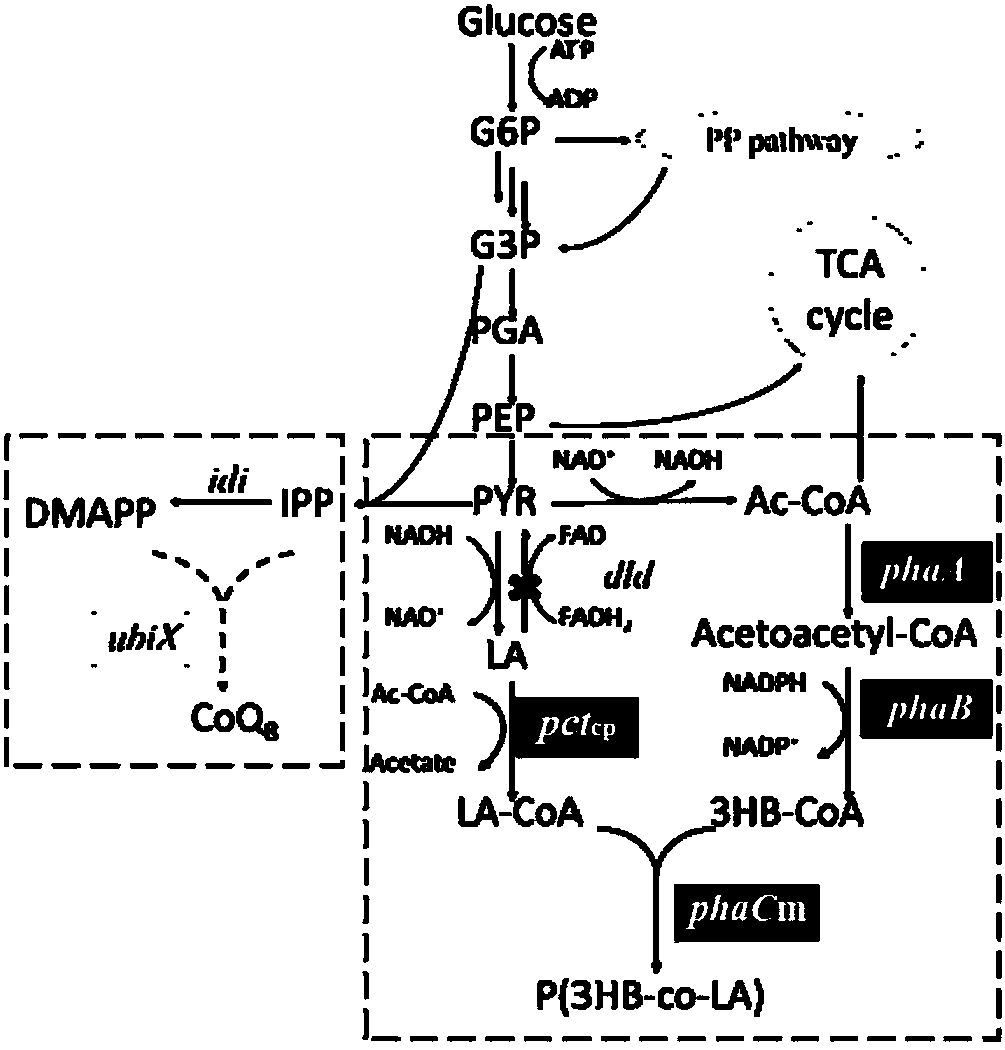
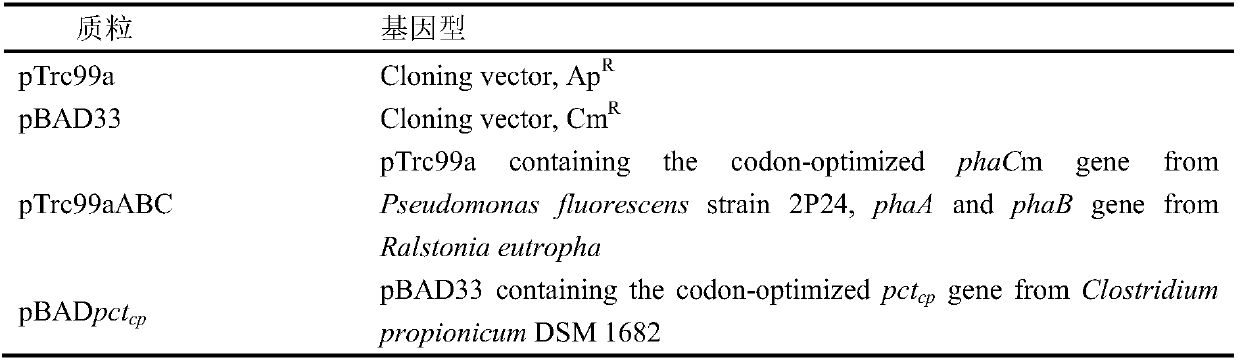
Method for improving the content of lactic acid composition in escherichia coli synthesized poly(3-hydroxybutyrate-co-lactic acid)
ActiveCN110295188AIncrease mole percentageIncrease the content of lactic acid componentsBacteriaMicroorganism based processesEscherichia coliHydroxybutyric acid
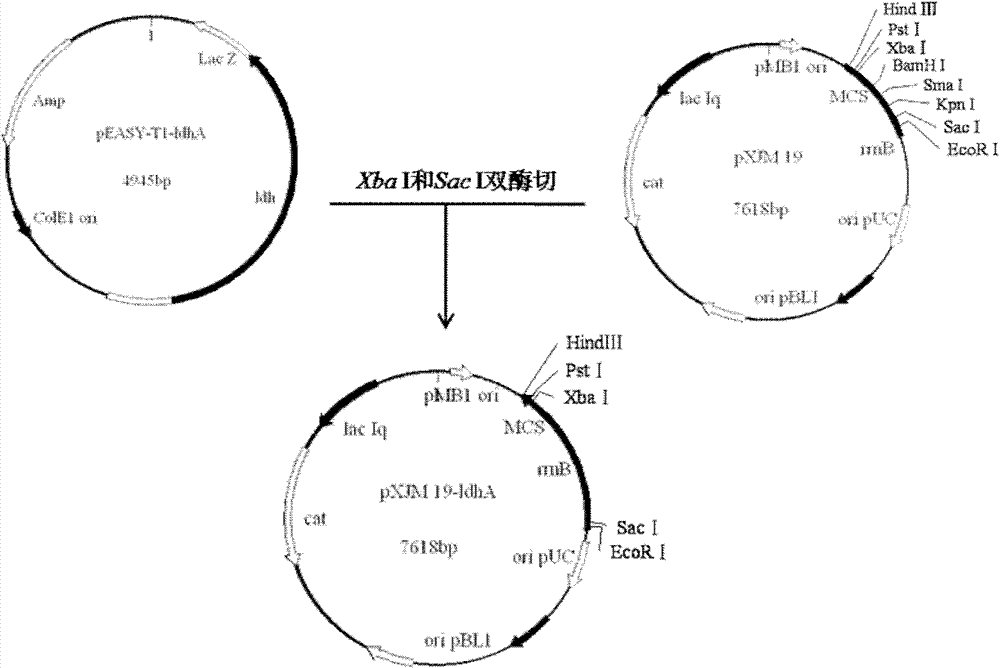
The invention provides a method for improving the content of the lactic acid component in escherichia coli synthesized poly(3-hydroxybutyrate-co-lactic acid). On the basis of recombinant escherichia coli E.coli MG-01, a flavin isopentenyl transferase gene ubiX is knocked out, and / or a D-lactic dehydrogenase lactate gene dld is knocked, and / or a constitutive ldhA promoter is replaced with a promoter expressing the propionyl coenzyme A transferase pctcp. Accordingly, by analyzing metabolic pathways and control, by means of the genetic engineering means, escherichia coli is transformed, and underthe aerobic condition, obtained strains can improve the mole percent of the lactic acid composition in the poly(3-hydroxybutyrate-co-lactic acid) synthesized in the escherichia coli.
Owner:EAST CHINA UNIV OF SCI & TECH
Engineered microorganism producing homo-succinic acid and method for preparing succinic acid using the same
ActiveUS9217138B2Increase productivityHigh rateBacteriaStable introduction of DNAHigh concentrationMannheimia
The present invention relates to a mutant microorganism, which is selected from the group consisting of genus Mannheimia, genus Actinobacillus and genus Anaerobiospirillum, producing homo-succinic acid and a method for producing homo-succinic acid using the same, and more particularly to a mutant microorganism producing succinic acid at a high concentration while producing little or no other organic acids in anaerobic conditions, which is obtained by disrupting a gene encoding lactate dehydrogenase (ldhA), a gene encoding phosphotransacetylase (pta), and a gene encoding acetate kinase (ackA), without disrupting a gene encoding pyruvate formate lyase (pfl), as well as a method for producing succinic acid using the same. The inventive mutant microorganism has the property of having a high growth rate and succinic acid productivity while producing little or no organic acids, as compared to the prior strains producing succinic acid. Thus, the inventive mutant microorganism is useful to produce succinic acid for industrial use.
Owner:KOREA ADVANCED INST OF SCI & TECH
High-D-lactic acid-yield recombinant bacterium and application thereof
InactiveCN106520644AEfficient synthesisHigh optical purityBacteriaMicroorganism based processesK pneumoniaeEconomic benefits
The invention discloses a high-D-lactic acid-yield recombinant bacterium and application thereof, belonging to the field of bioengineering. The recombinant bacterium provided by the invention is recombinant Klebsiella pneumoniae obtained by overexpressing D-lactate dehydrogenase gene ldhA in Klebsiella pneumoniae. According to the production method of D-lactic acid, progressive amplification culture is carried out after the recombinant bacterium is activated, thereby implementing pilot-scale production on the 500L fermentation tank level. The production method of D-lactic acid is simple and easy to implement, and implements high-efficiency synthesis of D-lactic acid on the pilot-scale level; the yield reaches 210 g / L, the D-lactic acid is free of L-lactic acid; and the recombinant bacterium has the characteristics of high yield and lower production cost, and has favorable market prospects and economic benefits.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
Biocatalyst for production of D-lactic acid
ActiveUS8669093B2High selectivityHigh optical puritySugar derivativesBacteriaEscherichia coliD-lactate dehydrogenase
A method for producing D-lactic acid in high yield, and to provide a method for producing D-lactic acid with high selectivity, in which optical purity is high and a by-product organic acid is small. In one aspect, a microorganism, wherein activity of pyruvate formate-lyase (pfl) is inactivated or decreased, and further activity of Escherichia coli-derived NADH-dependent D-lactate dehydrogenase (ldhA) is enhanced, is cultured to efficiently produce D-lactic acid. With regard to a method for enhancing ldhA activity, by linking, on a genome, a gene encoding ldhA with a promoter of a gene which controls expression of a protein involved in a glycolytic pathway, a nucleic acid biosynthesis pathway or an amino acid biosynthesis pathway, suitable results are obtained compared to the method for enhancing expression of the gene using an expression vector. A microorganism in which a dld gene is substantially inactivated or decreased is cultured to produce high quality D-lactic acid with reduced concentration of pyruvic acid.
Owner:MITSUI CHEM INC
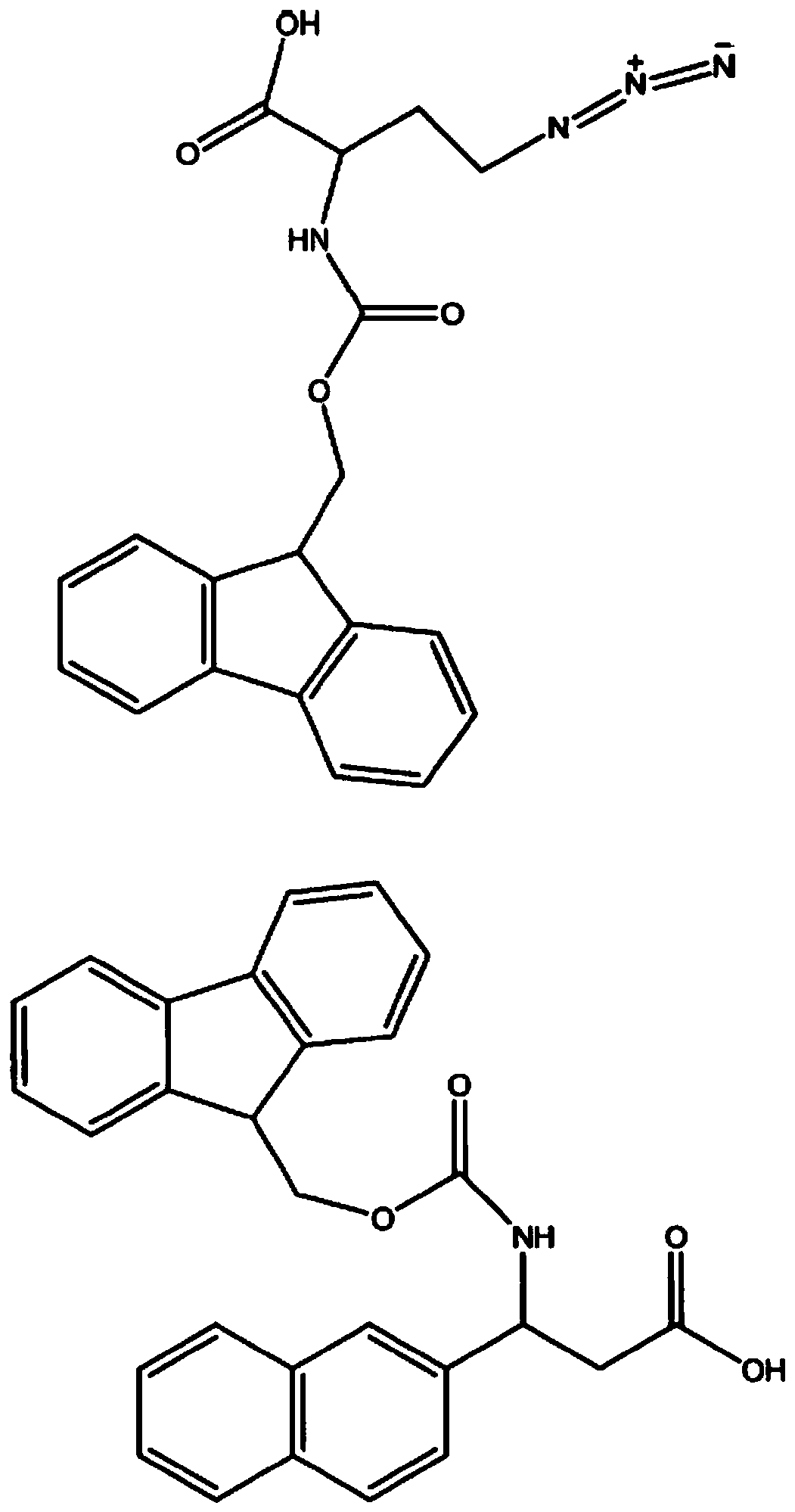
Synthesizing method of key intermediate for synthesizing lactate dehydrogenase A (LDHA) inhibitor
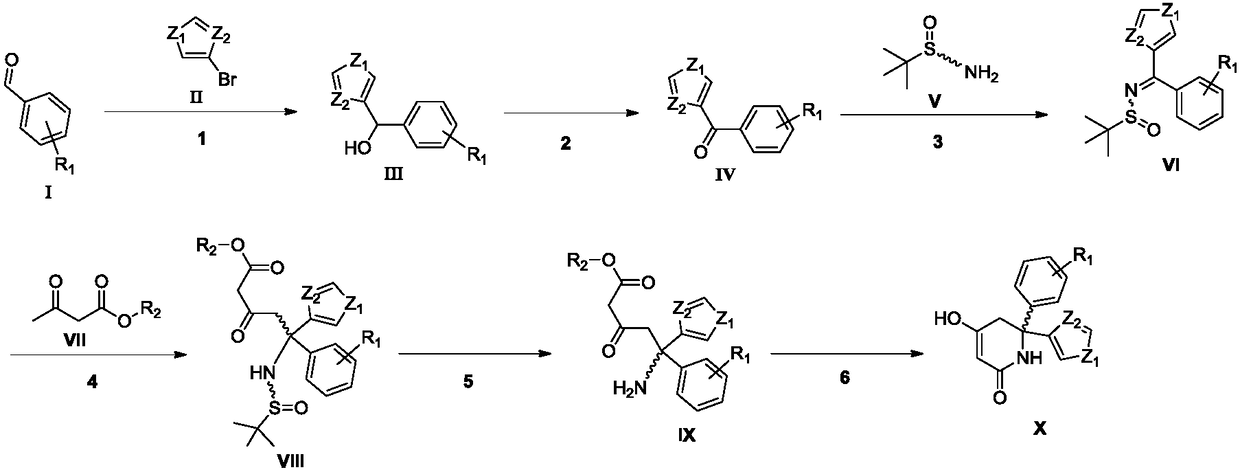
ActiveCN109369631AMild reaction conditionsAvoid low temperature reactionOrganic chemistry methodsAfter treatmentLactate dehydrogenase A
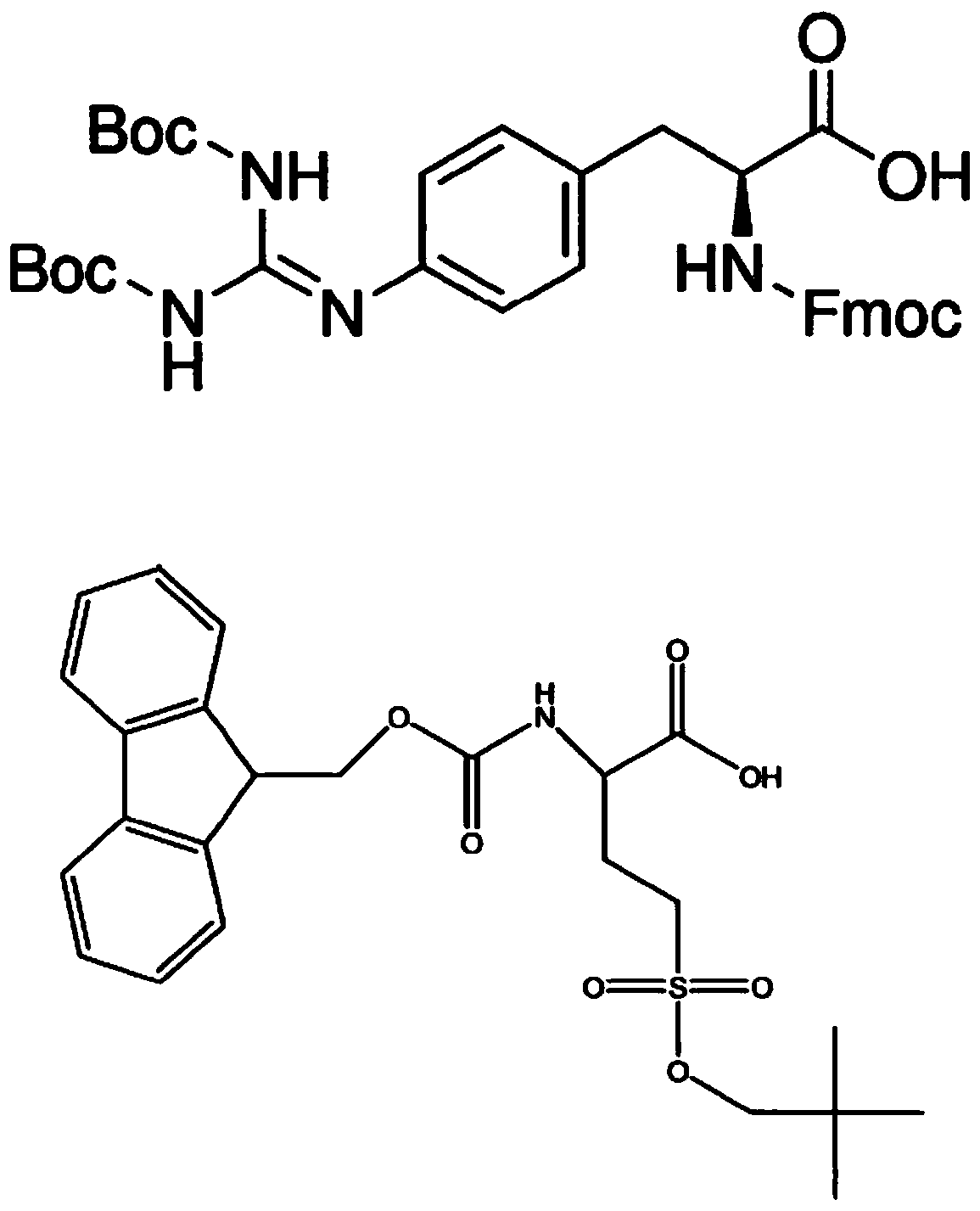
The invention relates to a synthesizing method of a key intermediate compound X for synthesizing a lactate dehydrogenase A (LDHA) inhibitor. According to the synthesizing method, a compound IX is prepared through the steps such as addition, oxidization, replacing and acidolysis by adopting a compound I as a raw material; and a the compound X is prepared by the compound IX through a enzyme ring closing, and through the method, the compound X with the single configuration can be prepared, wherein the ee value of the compound X is higher than 99%. A reaction is easy to operate, simple easy and convenient in subsequentto after-treatment, high in total yield, environmentally friendly, low in cost and suitable for industrial production. (Pplease see the specifications for the chemical structuralformulas).
Owner:SHANGHAI HAOYUAN MEDCHEMEXPRESS CO LTD
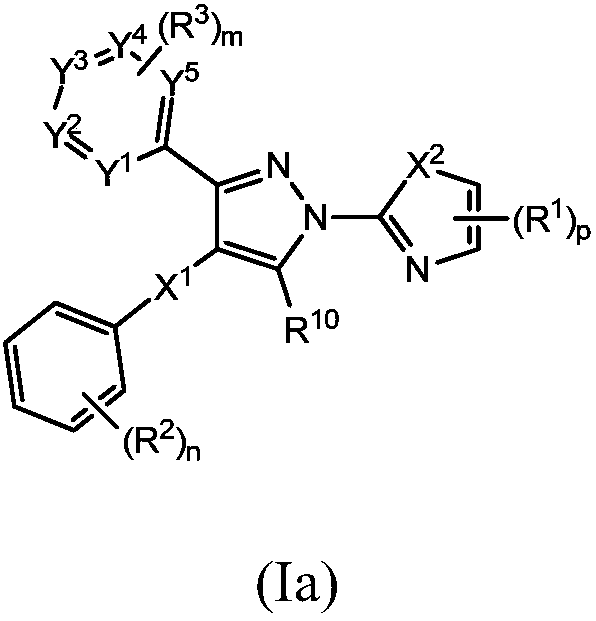
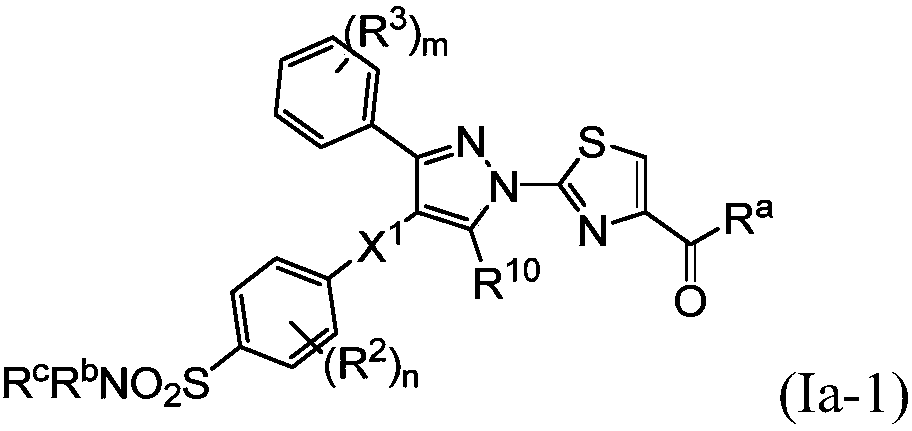
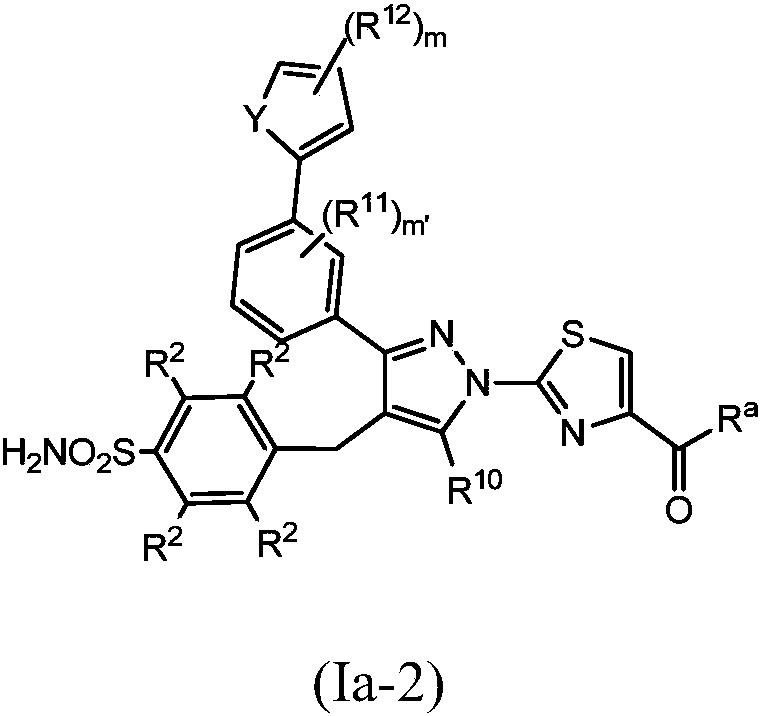
Small molecule inhibitors of lactate dehydrogenase and methods of use thereof
Provided is a compound of formula (I) [Formula (I) should be inserted here], in which Ar1, R1, U, V, W, X, and p are as described herein. Also provided are methods of using a compound of formula (I),including a method of treating cancer, a method of treating a patient with cancer cells resistant to an anti-cancer agent, and a method of inhibiting lactate dehydrogenase A (LDHA) and / or lactate dehydrogenase B (LDHB) activity in a cell.
Owner:US DEPT OF HEALTH & HUMAN SERVICES +3
Genetically engineered bacteria with high electroactivity and environmental stress tolerance
ActiveCN106520653BImprove electricity production activityHigh electroactivityBacteriaMicroorganism based processesSynechococcusBiotechnology
The invention relates to a genetically engineered bacterium with high-yield electroactivity and environmental stress tolerance. The lactic dehydrogenase gene ldhA in a pseudomonas aeruginosa PAO1 genome is knocked out, and the PAOl knockout strain is obtained; then a global regulatory factor IrrE of the deinococcus radiodurans is guided into the ldhA-, and the genetic engineering strain ldhA--irrE is obtained. After the thallus treated through a chemical agent is inoculated to a microbial fuel cell, the generated voltage and power density are improved by 46.76%-53.33% than those of the wild type strain, the time for stabilizing a system is shortened by 28.57%, the internal resistance of the system is reduced by 12.66%, the voltage under polyethylene glycol treatment is 543, the power density is 207, the stabilizing time is 130 h, the internal resistance reaches 420.32, and the survival rate of the engineered strain under the hunger condition, the high-acid-base condition and high-salt condition are improved by one time than those of the wild type strain.
Owner:TIANJIN UNIV OF SCI & TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com