A method for gene quantitative analysis
A quantitative analysis and gene technology, applied in the fields of biotechnology and medicine, can solve problems such as unrealistic, affecting the clarity of large charts, and inconsistent meanings of CT
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A method for quantitative analysis of single-cell housekeeping gene beta-Actin, comprising the following steps:
[0030] 1. Take a group of samples containing one or a small number of cells, treat the cells with proteolytic enzyme (Proteinase K), use CellsDirect qRT-PCR kit (Invitrogen, USA) to synthesize cDNA, and pre-amplify the relevant fragments of beta-Actin gene 18cyces, used as beta-Actin gene quantification.
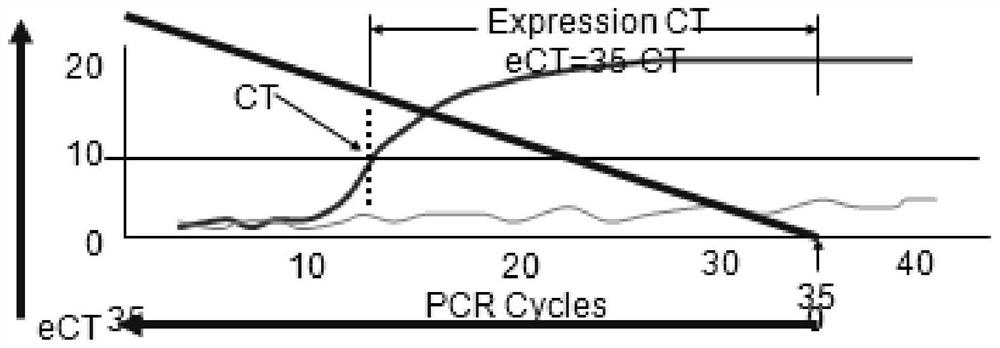
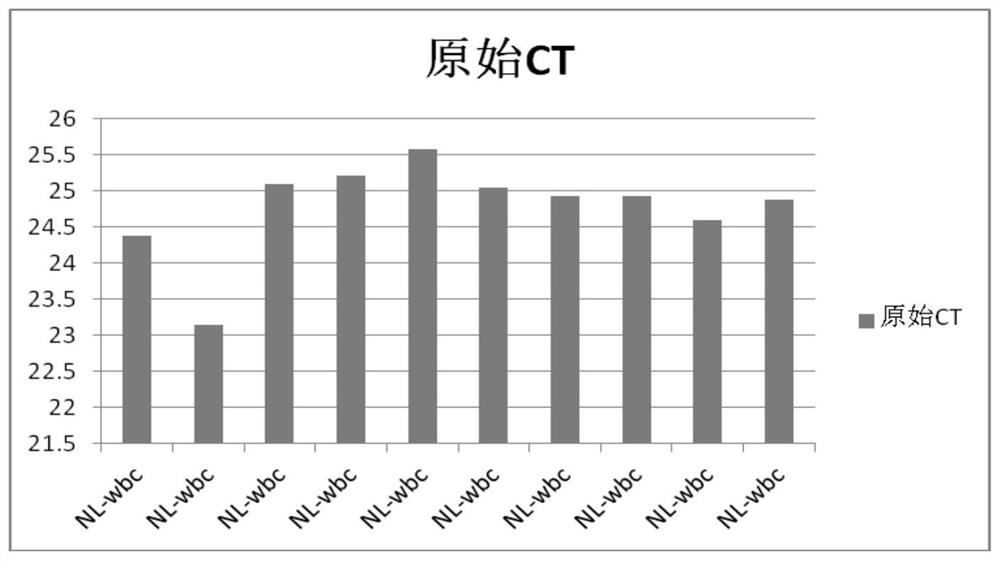
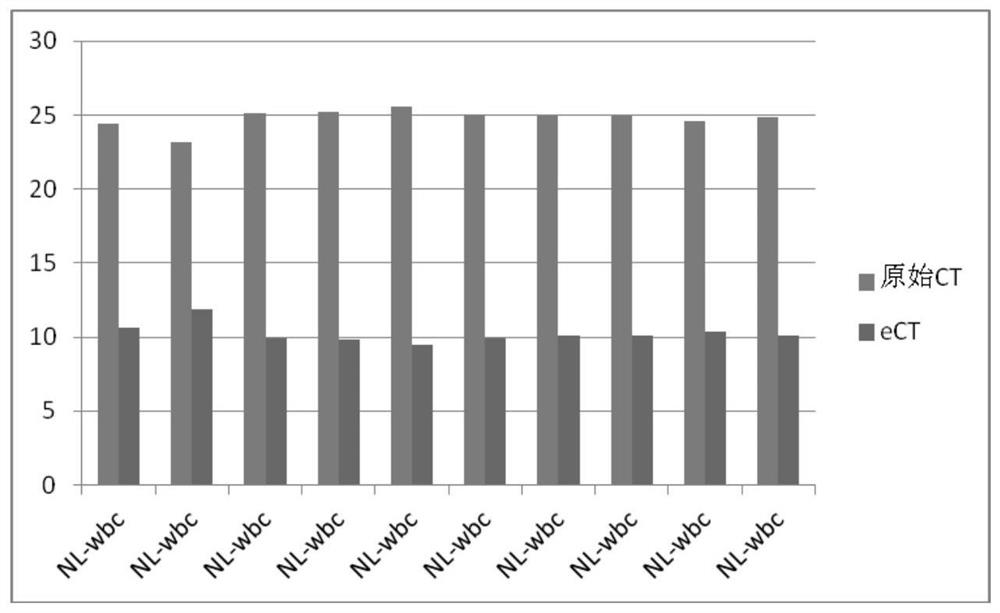
[0031] 2. With the gene template pre-amplified in step 1, use quantitative PCR equipment and reagents to detect the initial copy number of the beta-Actin gene in the pre-amplified sample, and obtain the original CT data, that is, the CT value, as shown in the table 1.
[0032] Table 1 Single-cell analysis of housekeeping gene beta-Actin
[0033] leukocyte Raw CT NL-wbc 24.37 NL-wbc 23.15 NL-wbc 25.1 NL-wbc 25.21 NL-wbc 25.57 NL-wbc 25.05 NL-wbc 24.92 NL-wbc 24.93 NL-wbc 24.6 NL-wbc 2...
Embodiment 2
[0043] A method for quantitative analysis of the single-cell level of single-cell housekeeping genes GAPDH, CD45, EpCAM, CK7, CK8, CK18, and CK19 genes, comprising the following steps:
[0044] 1. Take a group of samples containing one or a small number of cells, treat the cells with proteolytic enzyme (Proteinase K), use CellsDirect qRT-PCR kit (Invitrogen, USA) to synthesize cDNA, and pre-amplify GAPDH, CD45, EpCAM, CK7 , CK8, CK18, and CK19 gene related fragments 18cyces, the gene template obtained by pre-amplification is used as the subsequent quantitative PCR;
[0045] 2. Use the gene template pre-amplified in step 1 for quantitative PCR test, and use quantitative PCR instruments and reagents to detect the initial copy number of multiple genes GAPDH, CD45, EpCAM, CK7, CK8, CK18, and CK19 in the same sample , to obtain the original CT data, that is, the CT value, as shown in Table 3.
[0046] Table 3 Raw CT values at the single-cell level of GAPDH, CD45, EpCAM, CK7, CK8...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com