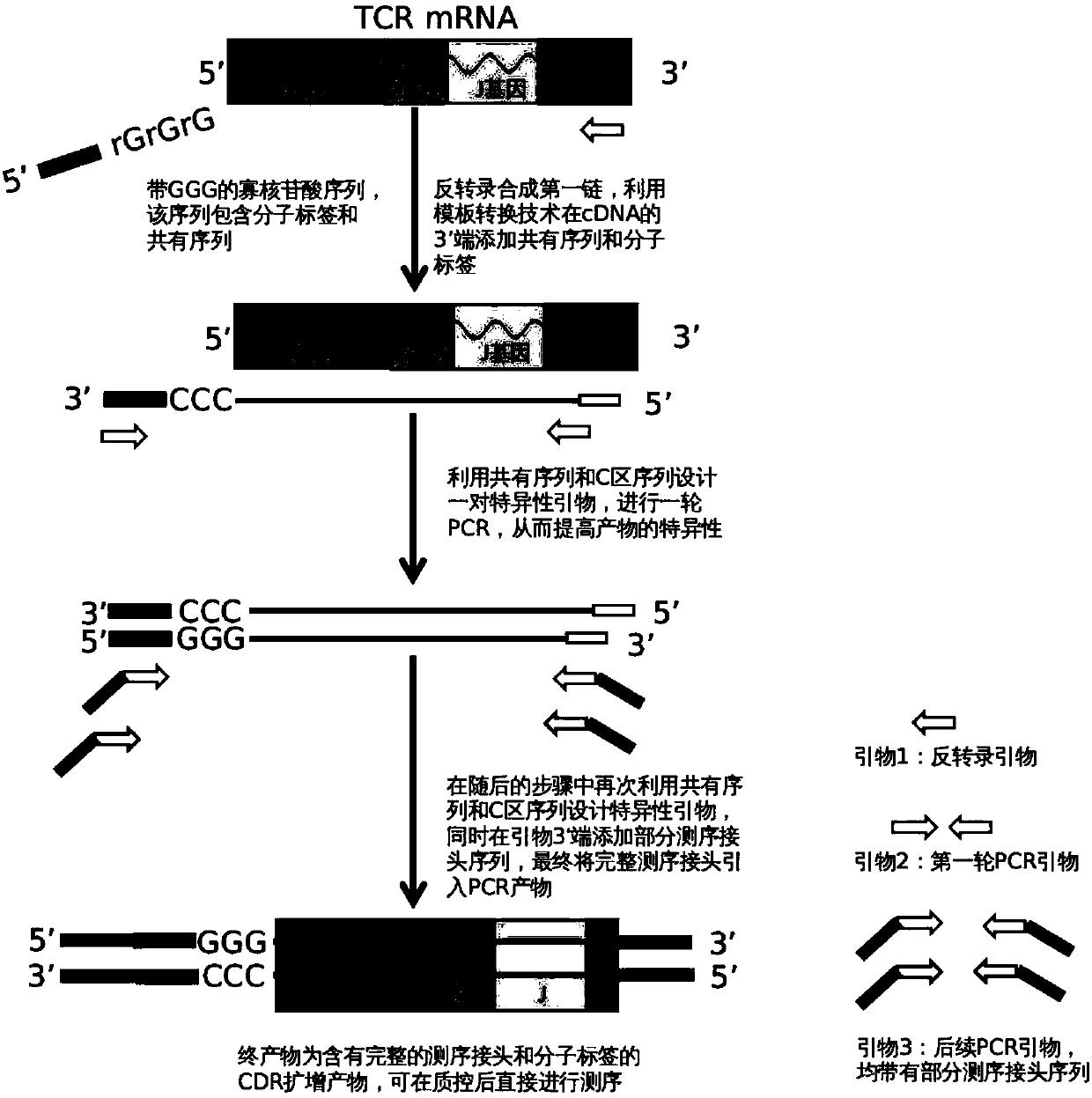
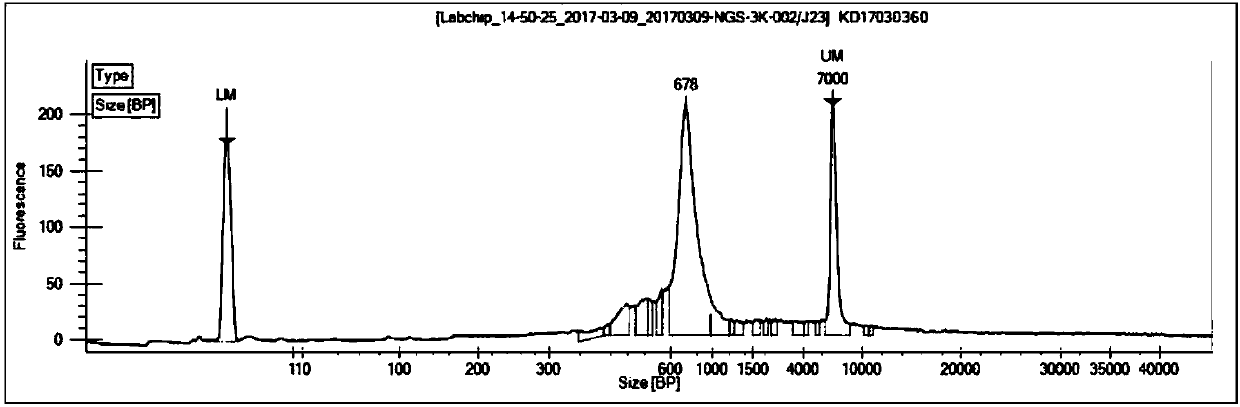
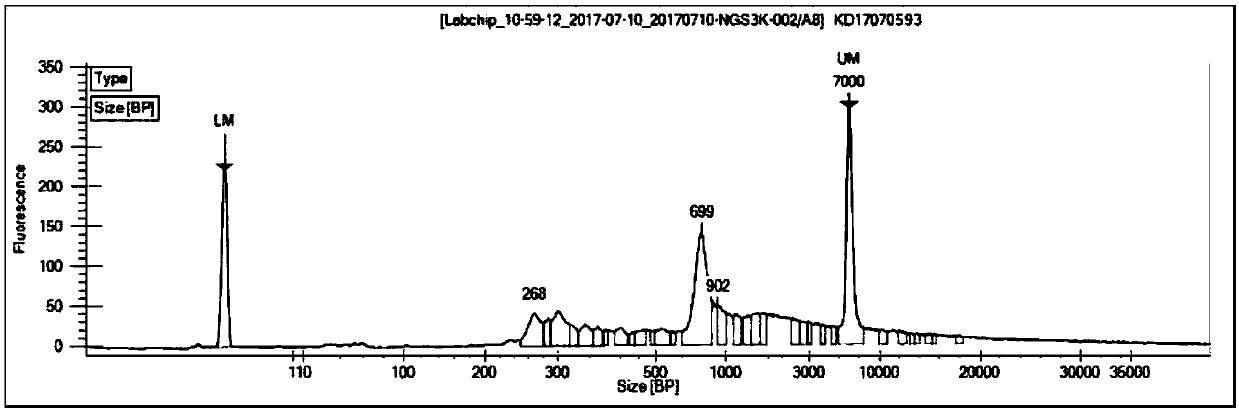
Construction method of T cell antigen receptor diversity sequencing library and kit
A technology for sequencing libraries and kits, used in chemical libraries, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems of difficult primer design, uneconomical, and biased results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0131] 1. Experimental primers:
[0132] Table 1:
[0133]
[0134]
[0135] All the primers in Table 1 above were dissolved and made into a stock solution with a concentration of 100 μM. The working solution concentration is 10 μM.
[0136] 2. Experimental process:
[0137] 2.1 Total RNA extraction: total RNA was extracted from mouse spleen as a biological sample.
[0138] 2.2 Reverse transcription: TRBC-RT and 5TSA in the above table 1 are used as primers, using SuperScript TM II ReverseTranscriptase (Thermo Fisher#18064014) kit, perform primer-specific reverse transcription to generate cDNA template, the specific steps are as follows:
[0139] (1) RNA denaturation, incubation with primers:
[0140] Adjust the RNA concentration to 100-1000 ng / μL, take 2 μL, add 1 μL of TRBC-RT (10 μM), mix well, place in a 70°C water bath for 2 minutes, then place in a 42°C water bath for 2 minutes;
[0141] (2) Reverse transcription, through template replacement, introduce molec...
Embodiment 2
[0174] 1. Experimental primers:
[0175] Table 4:
[0176]
[0177]
[0178] All the primers in Table 1 above were dissolved and made into a stock solution with a concentration of 100 μM. The working solution concentration is 10 μM.
[0179] 2. Experimental process:
[0180] 2.1 Extraction of total RNA: 3 cases of human peripheral blood samples were used to separate white blood cells as biological samples, and total RNA was extracted.
[0181] 2.2 Reverse transcription: TRBC-RT and 5TSA in the above table 1 are used as primers, using SuperScript TM II ReverseTranscriptase (Thermo Fisher#18064014) kit, perform primer-specific reverse transcription to generate cDNA template, the specific steps are as follows:
[0182] (1) RNA denaturation, incubation with primers:
[0183] Take 700ng RNA, add 1 μL of TRBC-RT (10 μM), add water to 6.9 μl, mix well, and put in a PCR machine at 70°C for 2 minutes, and at 42°C for 2 minutes;
[0184] (2) Reverse transcription, through te...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com