Application of lncRNA in diagnosis and prognostic prediction of liver cancer
A technology of liver cancer, CTD-2574D22.4, applied in the field of biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
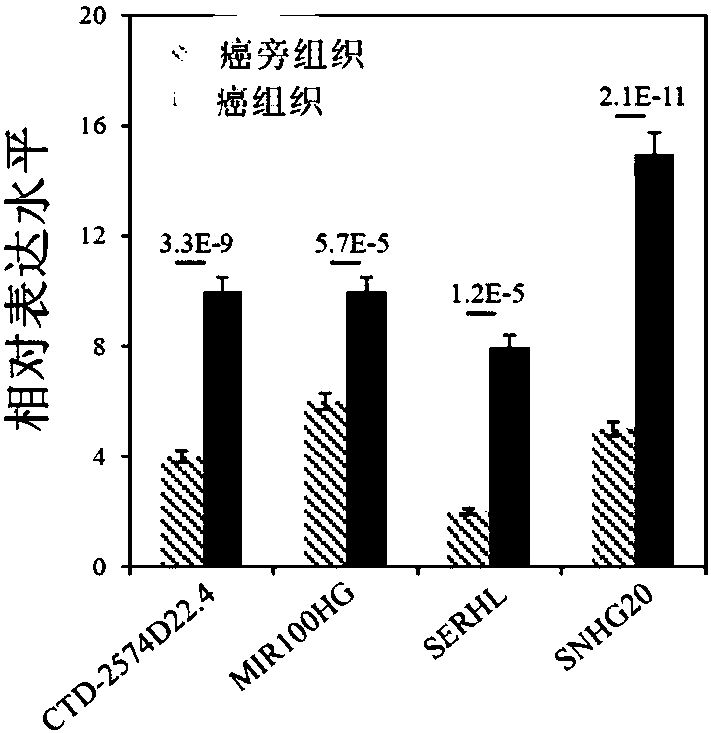
[0046] Example 1 QPCR detection of lncRNA expression level in patients with liver cancer
[0047] 1. Sample collection
[0048] The cancer tissues and paracancerous tissues of 84 liver cancer patients were collected, and the patients gave informed consent, and all the above specimens were obtained with the consent of the organizational ethics committee.
[0049] 2. RNA extraction
[0050] The tissue RNA extraction kit of Invitrogen was used to extract the tissue RNA, and the operation was performed according to the specific steps in the manual.
[0051] 3. Reverse transcription:
[0052] Use 25μl reaction system, take 1μg total RNA for each sample as template RNA, and add the following components to PCR tubes: DEPC water, 5× reverse transcription buffer, 10mM dNTP, 0.1mM DTT, 30μM Oligo dT, 200U / μl M-MLV, template RNA. Incubate at 42°C for 1 hour, then centrifuge briefly at 72°C for 10 minutes.
[0053] QPCR amplification test
[0054] Primers were designed according to ...
Embodiment 2
[0078] Example 2 Detection of the impact of IncRNA on the survival period of patients with liver cancer
[0079] 1. Sample
[0080]From the TCGA database, 364 cases of liver cancer tissues and paracancerous tissues of consecutive pathologically diagnosed and surgically resected liver cancer cases were screened and randomly divided into a training group (182 cases) and a verification group (182 cases). The clinical information of the included patients included age. , gender, tumor grade, tumor stage, new tumors, and survival information.
[0081] 2. Data analysis
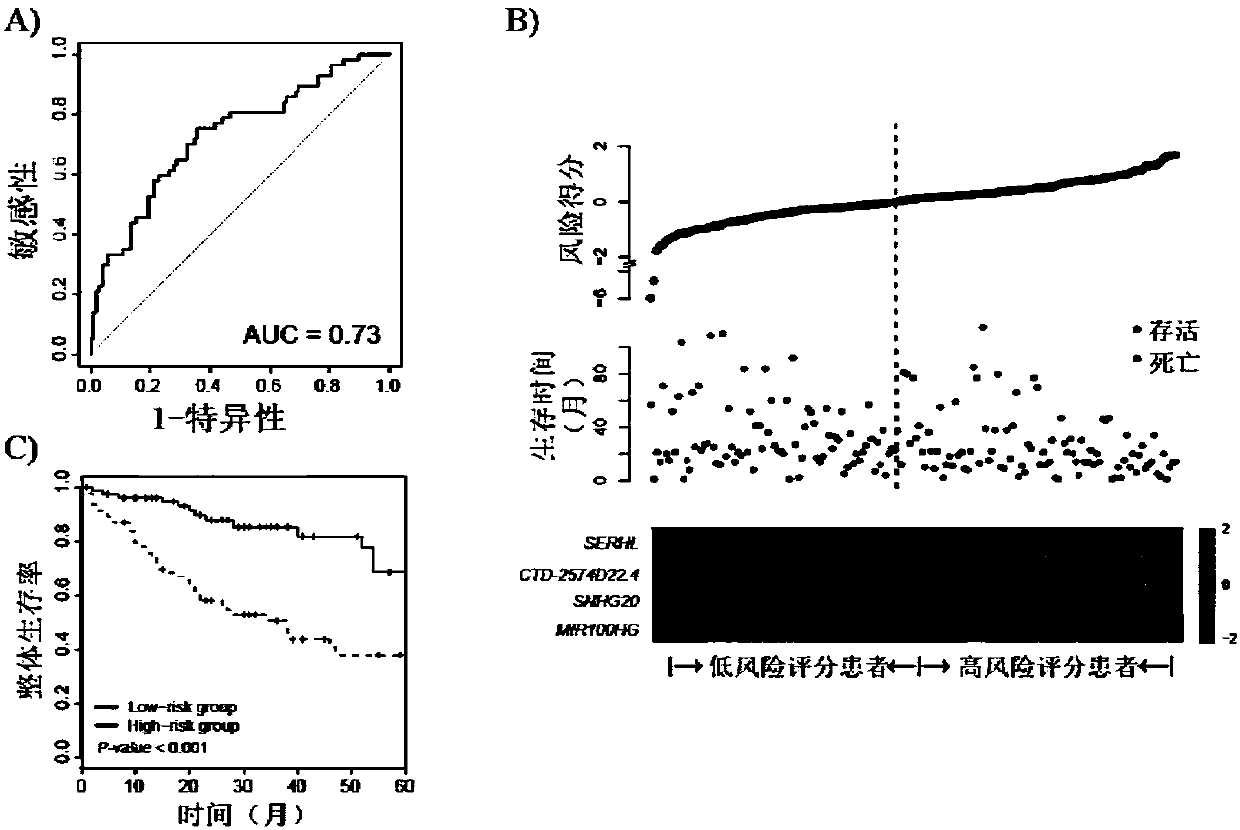
[0082] The lncRNA expression and survival time data were extracted, and the coxph function of the survival package was used to perform univariate Cox regression analysis on the lncRNA data of the training group, and the p value in the univariate Cox regression was screened to obtain a p value <0.01.
[0083] 3. Results
[0084] Through univariate Cox regression analysis, it was found that differentially expressed ...
Embodiment 3
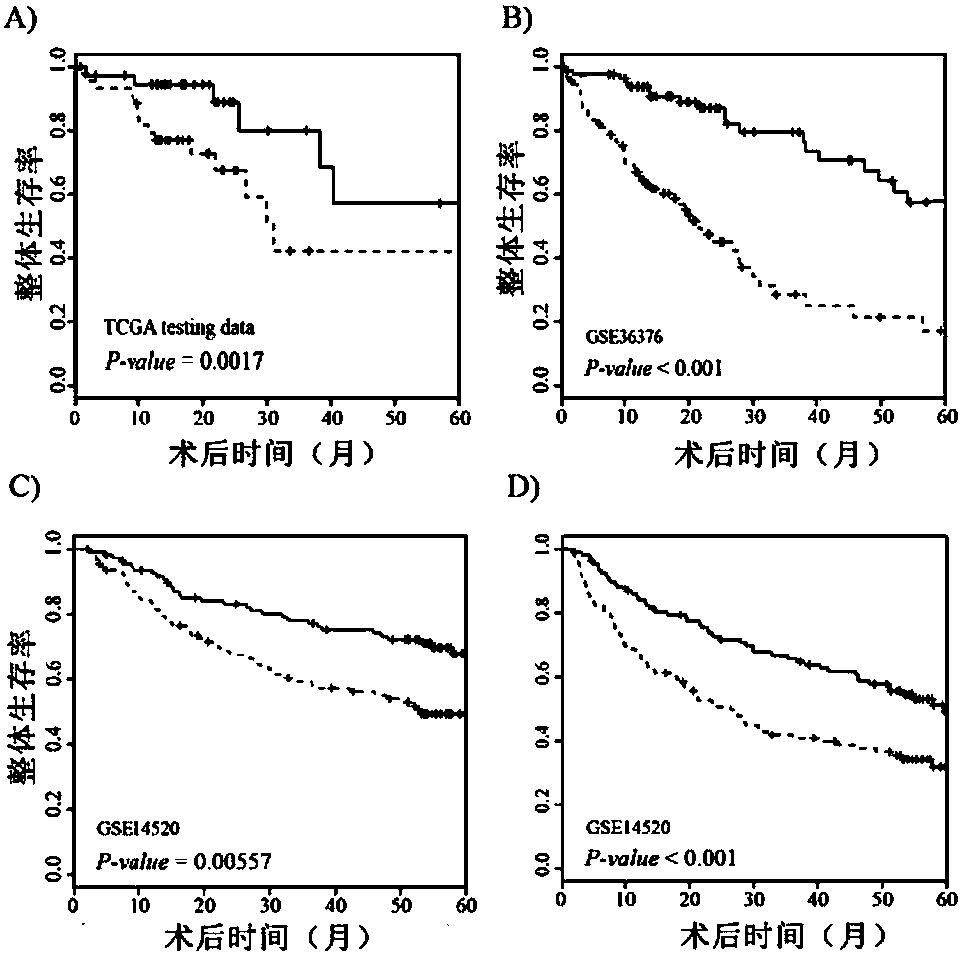
[0085] Example 3 Efficiency Test of RS Model in Predicting the Prognosis of Liver Cancer Patients
[0086] 1. Data analysis
[0087] The prognostic features of each screened lncRNA were analyzed by univariate Cox regression. In the training group, lncRNAs were associated with the overall survival of patients (P<0.01). The contribution of each lncRNA to survival prediction was then calculated using multivariate Cox regression in the R package. A risk scoring model is established to assess the risk of patients, and the scoring formula is as follows:
[0088] risk score Where N represents the number of lncRNAs used for prognosis prediction, Expi represents the expression level of lncRNAi, and Ci represents the regression coefficient of lncRNAi obtained from multivariate Cox regression analysis.
[0089] ROC curves and AUC curves were used to determine the sensitivity and specificity of RS model predictions. Kaplan-Meier survival curves were used to analyze the difference bet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com