Method and device for detecting gene fusion of target areas and storage medium
A gene fusion, target area technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of insufficient data consideration, sensitivity defects, inability to maximize computing resources and running time, etc., to achieve optimization Resource requirements and detection speed, effects of improved sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
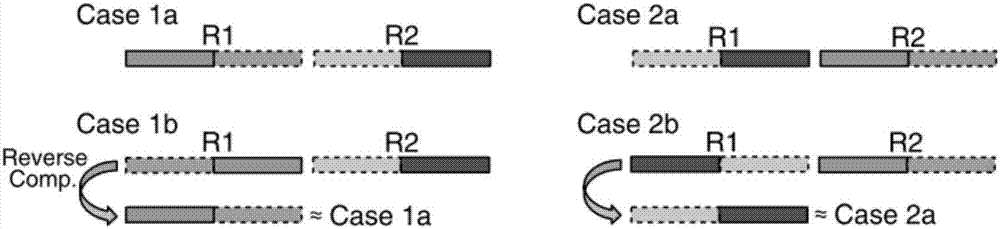
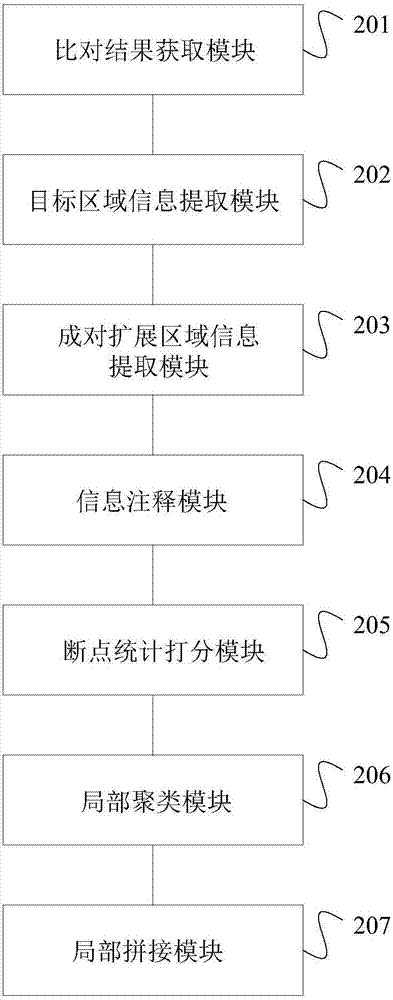
[0065] according to figure 1 In the process shown, BWA is used to compare the PE sequencing data of the target region of the quality-assessed gene fusion positive sample (ALK exon 20-EML4exon13, 45-55%) with the human reference genome, after marking duplicated reads (duplicated reads) Wait for the processing process to obtain the comparison result in BAM format, and then use it as the input data of the method of the present invention to detect the gene fusion in the target region. Specifically, the method steps and detection parameter settings are as follows:
[0066] Obtain the result of comparing the paired-end sequencing data of the target region to the human reference genome; filter out sequences with multiple indels (InDel) or short tandem repeats; extract the abnormal size of the insert within the target region and within 200 bp ( Valid information (including alignment position, sequence base, quality value and alignment) of high-quality (quality value higher than 20) u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com