Oncolytic tumor viruses and methods of use
An adenovirus, tumor cell technology, applied in oncolytic tumor virus and its application field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
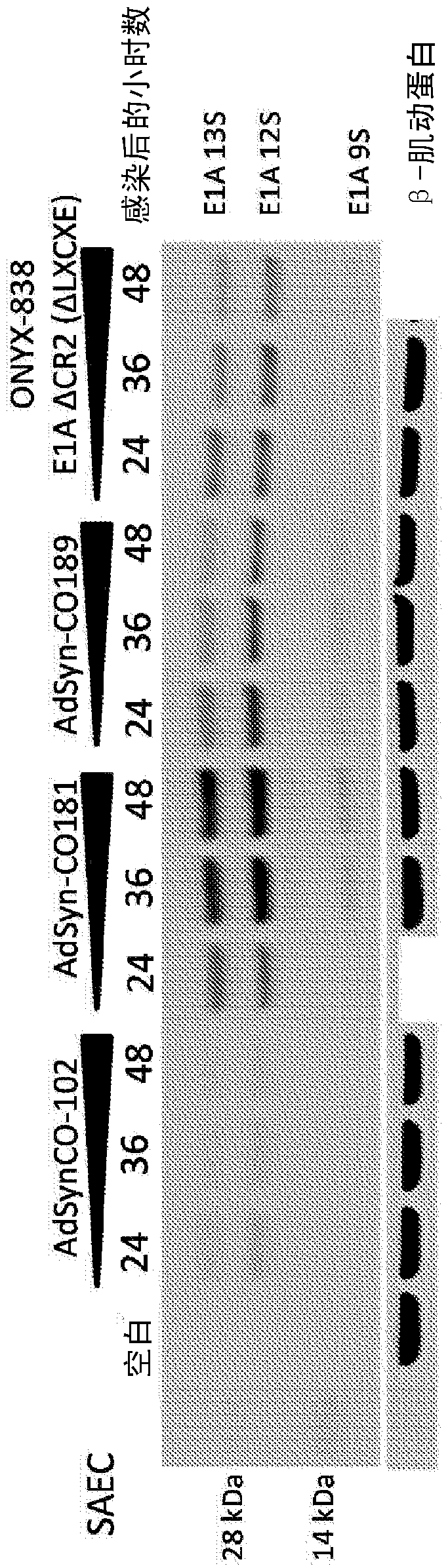
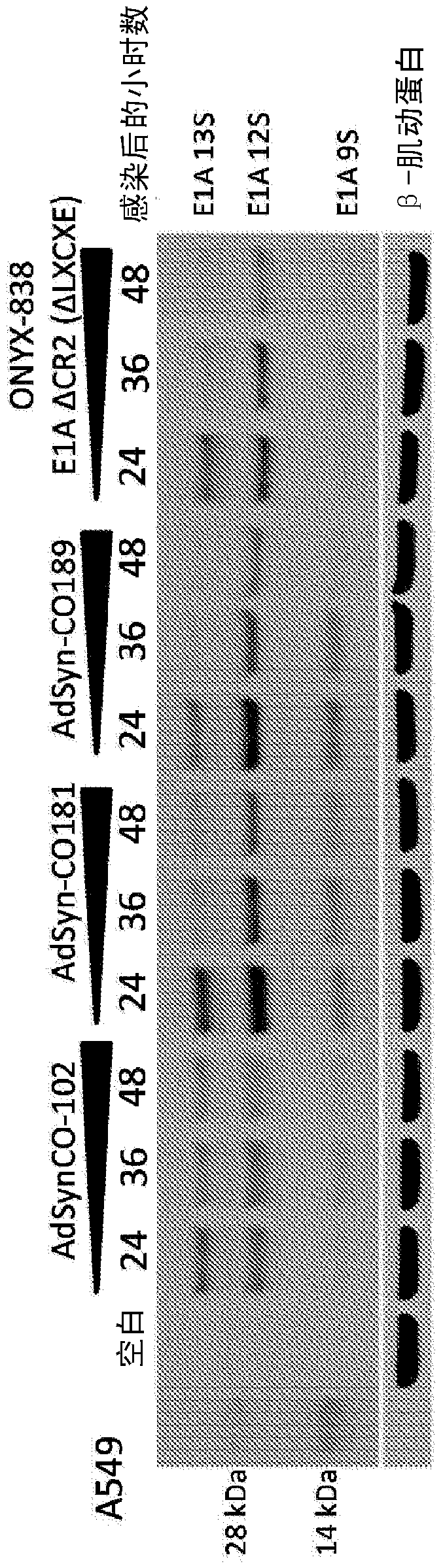
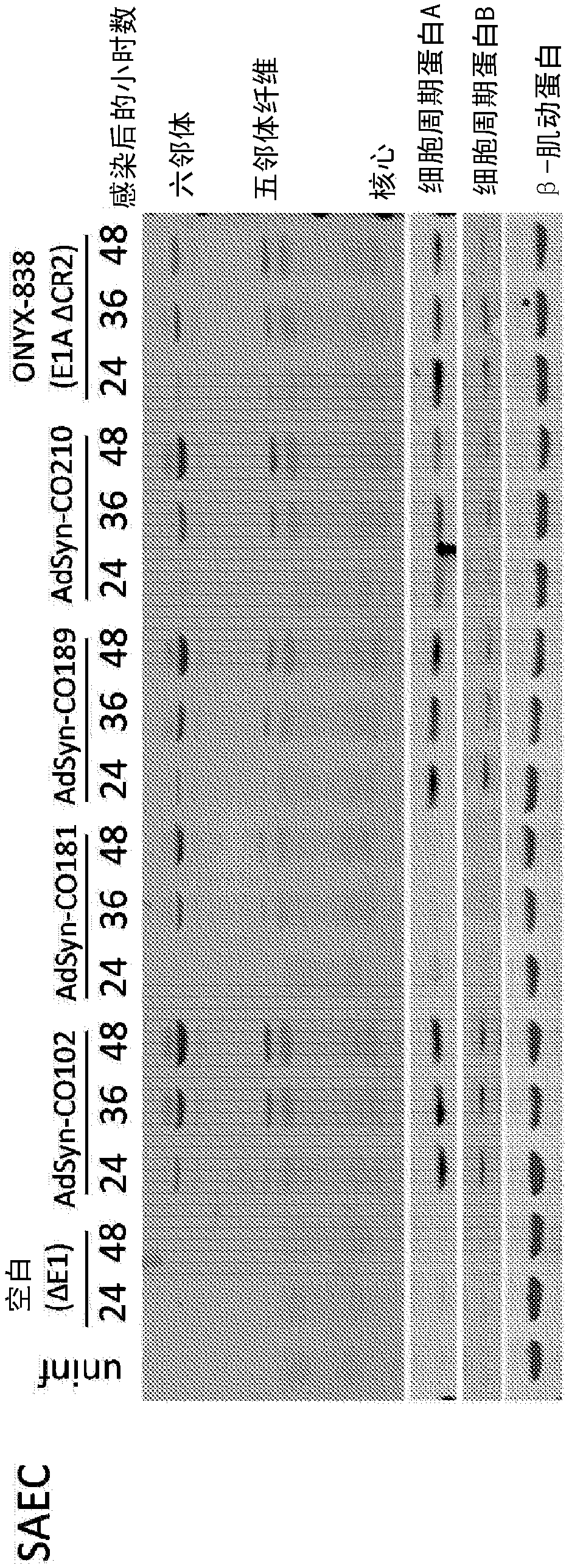
[0343] Example 1: Oncolytic Adenoviruses Selectively Replicate in Tumor Cells with Dysregulated E2F Activity
[0344] Modified adenoviruses were prepared from the components mentioned below according to the methods described in PCT Publication No. WO 2012 / 024351, incorporated herein by reference. The Gateway pDONR vector was used. The El module was obtained from human Ad5 DNA by PCR and inserted into the vector pDONR P1P4 using SLIC. The pDONR P1P4 vector backbone including attL1 and attL4 recombination sites was amplified using PCR and combined with the Ad5 E1 module using SLIC. The E3 module was obtained by PCR to generate a product flanked by attB5 and attB3r recombination sites. The product was inserted into pDONR P5P3r vector by Gateway BP reaction. The E4 module was obtained by PCR to generate a product flanked by attB3 and attB2 recombination sites. The product was inserted into pDONR P3P2 vector by Gateway BP reaction. The attR5-ccdB-Cm(r)-attR2 fragment from the ...
Embodiment 2
[0377]Example 2: Oncolytic Adenoviruses with Modifications in El, L3, E3 and / or E4
[0378] This example describes other recombinant adenoviruses with tumor selectivity. Exemplary recombinant viruses are listed in Table 2 and the genome sequences of these viruses are shown in SEQ ID NO: 25-31.
[0379] Table 2. Adenoviruses with modifications in E1, L3, E3 and / or E4
[0380]
[0381] AdSyn-CO312, AdSyn-CO313, and AdSyn-CO335 are rapamycin- and / or rapamycin analog (AP21967)-dependent EFGR-targeting viruses (see PCT Publication No. WO2013 / 138505, for which targeting The description of the method is hereby incorporated by reference in its entirety). Each of these recombinant viruses included deletions of the E3-RIDα / β and E3-14.7k coding sequences, as well as EGFRVHH-FKBP fusion proteins and FRB*-fibrils (Ad5 fibers fused to mutant FRB, which can bind rapamycin or rapamycin insertion of rapamycin analogues) or FRB-fibers (Ad5 fibers fused to wild-type FRB, which can only bi...
Embodiment 3
[0388] Example 3: Recombinant Adenovirus Expressing Chimeric Fiber Protein
[0389] This example describes recombinant adenoviruses expressing chimeric fibril proteins directed to infect specific cell types.
[0390] While Ad5 and many other serotypes have been shown to bind to the coxsackie adenovirus receptor (CAR) for cell attachment, other serotypes have been shown to use CD46, desmoglein 2, sialic acid, or others. Since Adsembly / AdSLIC (see PCT Publication No. WO 2012 / 024351, incorporated herein by reference) allows rapid generation of chimeric viruses, an initial set of six fiber chimeric viruses was generated to examine alternate cellular targets of various serotypes ability. Since the spherical nodule at the C-terminus of fibrin is generally responsible for receptor binding, chimeras were generated by replacing the Ad5 fibrous nodule with those from Ad3, Ad9, Ad11, Ad12, or Ad34 (FIGS. 23A23C; Table 3). Each virus generated had the same E1 module containing an E1A / E1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com