A method for identifying protein methylation
An identification method, methylation technology, applied in the field of identification of methylated proteomes, can solve the problems of inability to distinguish peptide segments, and achieve the effect of demethylation processing of spectral peaks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
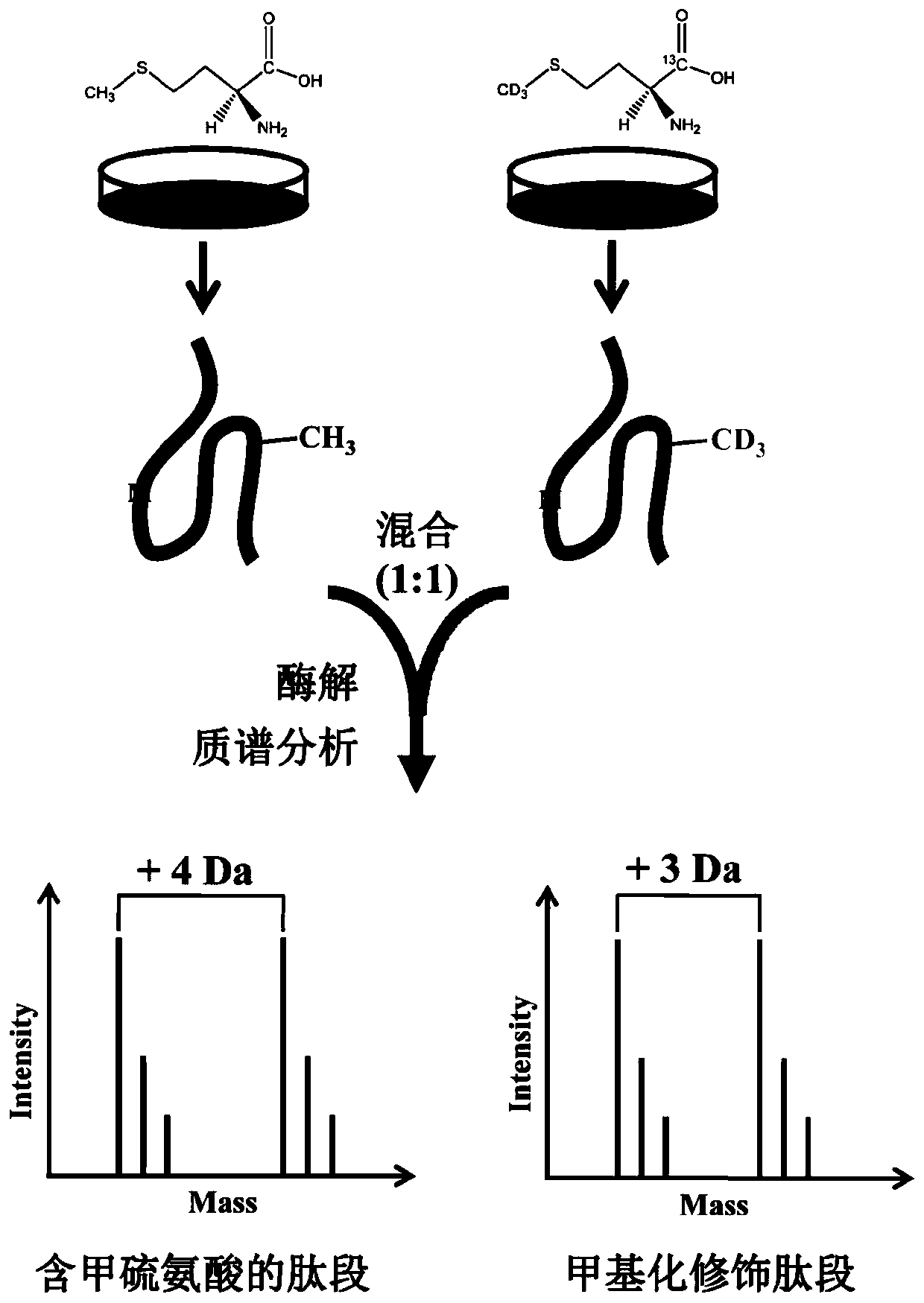
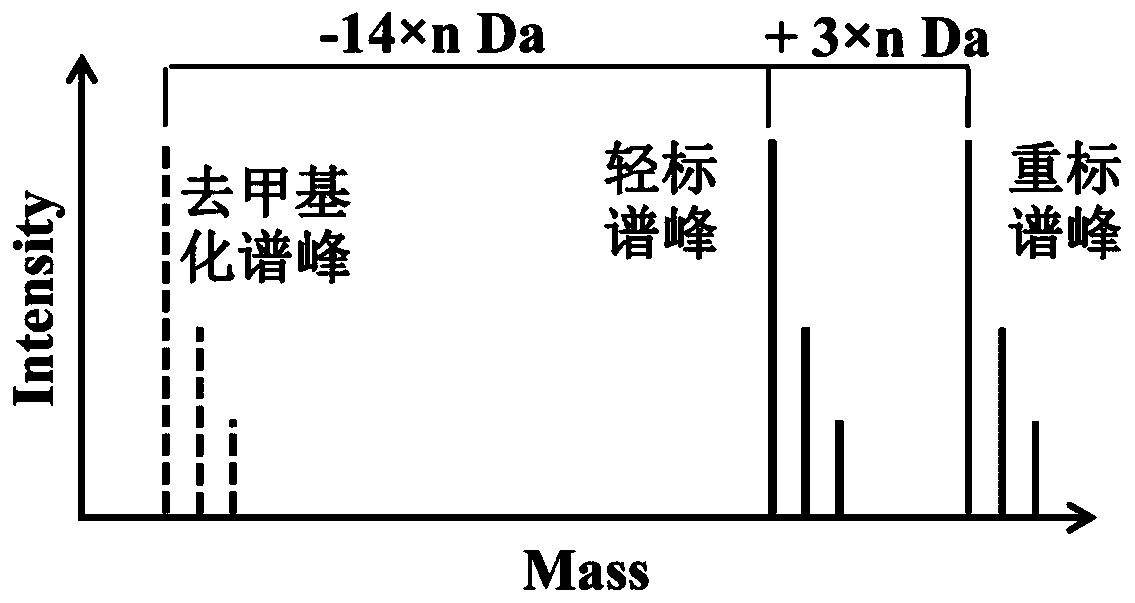
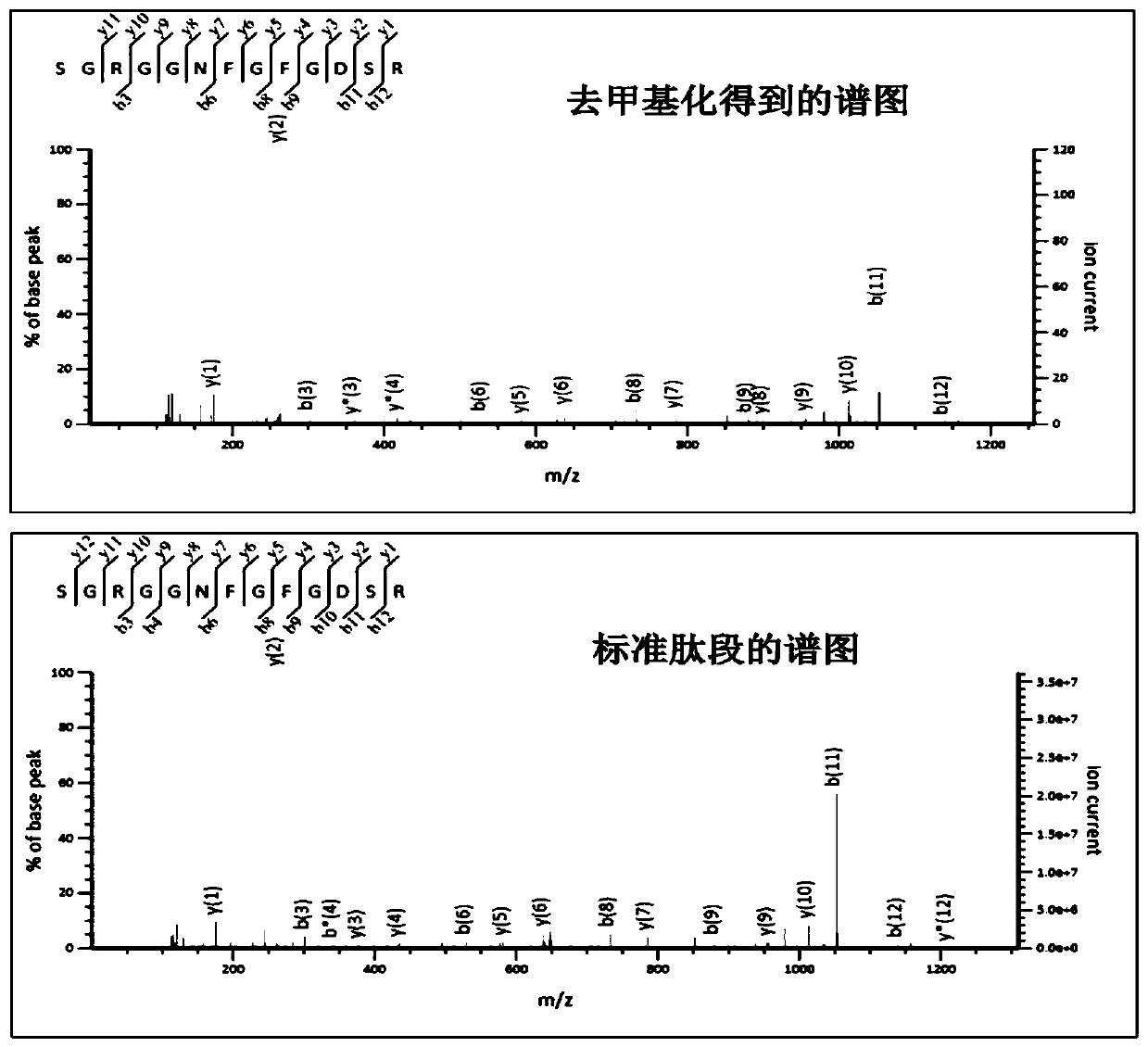
[0027] Peak demethylation strategy for large-scale analysis of methylation proteomics: HEK293 cells were cultured in DMEM containing 0.2 mM L‐methionine supplemented with 10% dialyzed fetal bovine serum Culture medium for eight generations (at least eight generations), in the presence of 0.2mM L-methionine-carboxyl- 13 C, Methyl‐D 3 Eight generations (at least eight generations) were cultured in the DMEM medium that added 10% of the fetal bovine serum of the dialyzed treatment; Collect the cells respectively and respectively in the lysate (8M urea, 1% v / v polyethylene glycol octyl phenyl ether , 65mM dithiothreitol, 1mM phenylmethylsulfonyl fluoride, 1% v / v protease inhibitor, 50mM Tris-Cl pH 7.5 buffer solution) in ultrasonic-assisted disruption; Samples and relabeled samples were mixed. Add a final concentration of 20 mM dithiothreitol to 2 mg of protein mixed sample, place in a water bath at 37 °C for 2 h, then add a final concentration of 40 mM iodoacetamide, react in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com