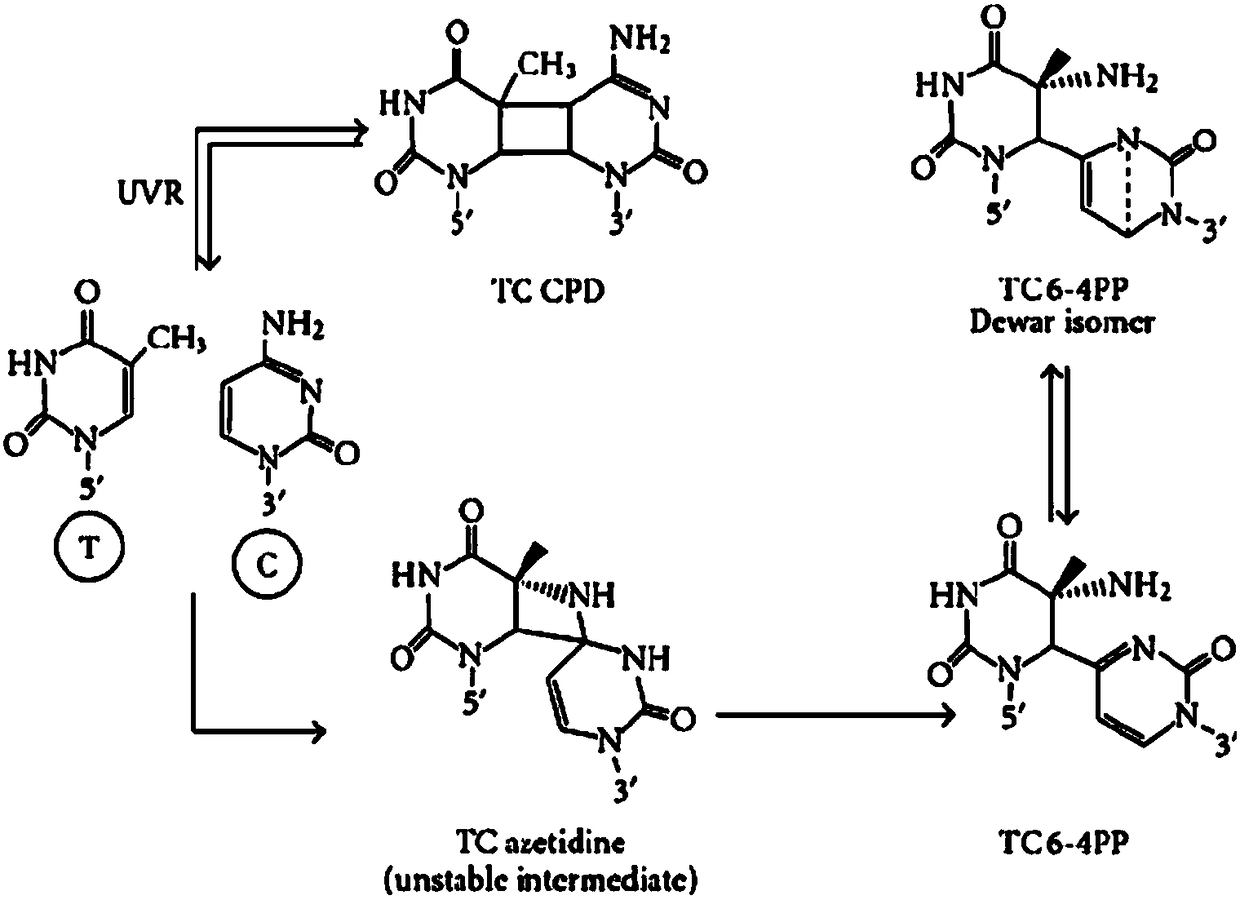
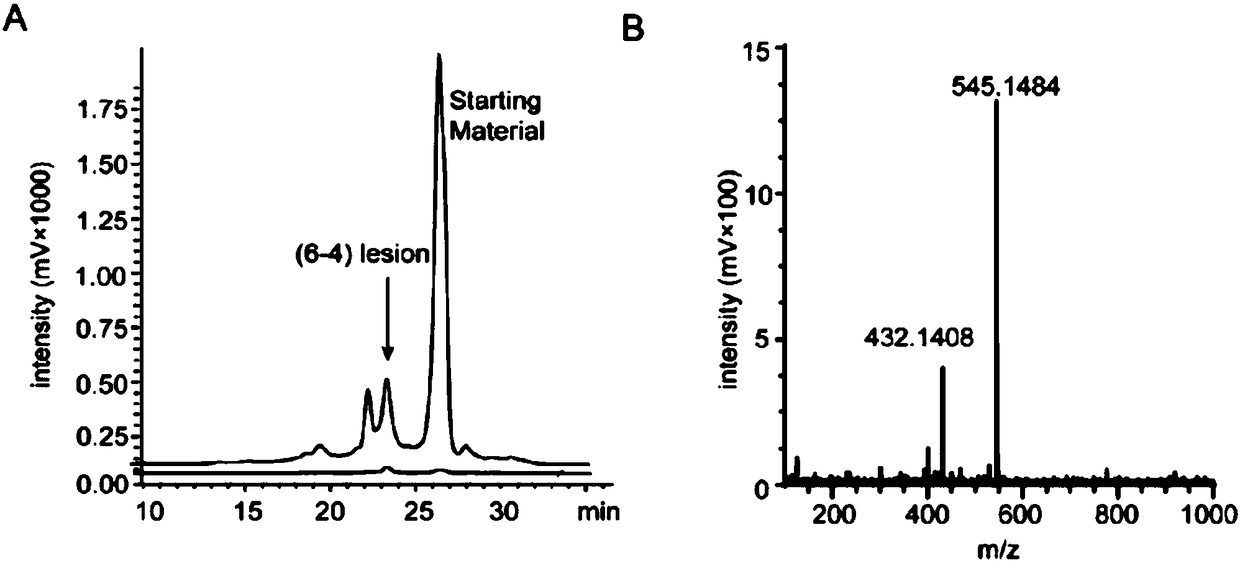
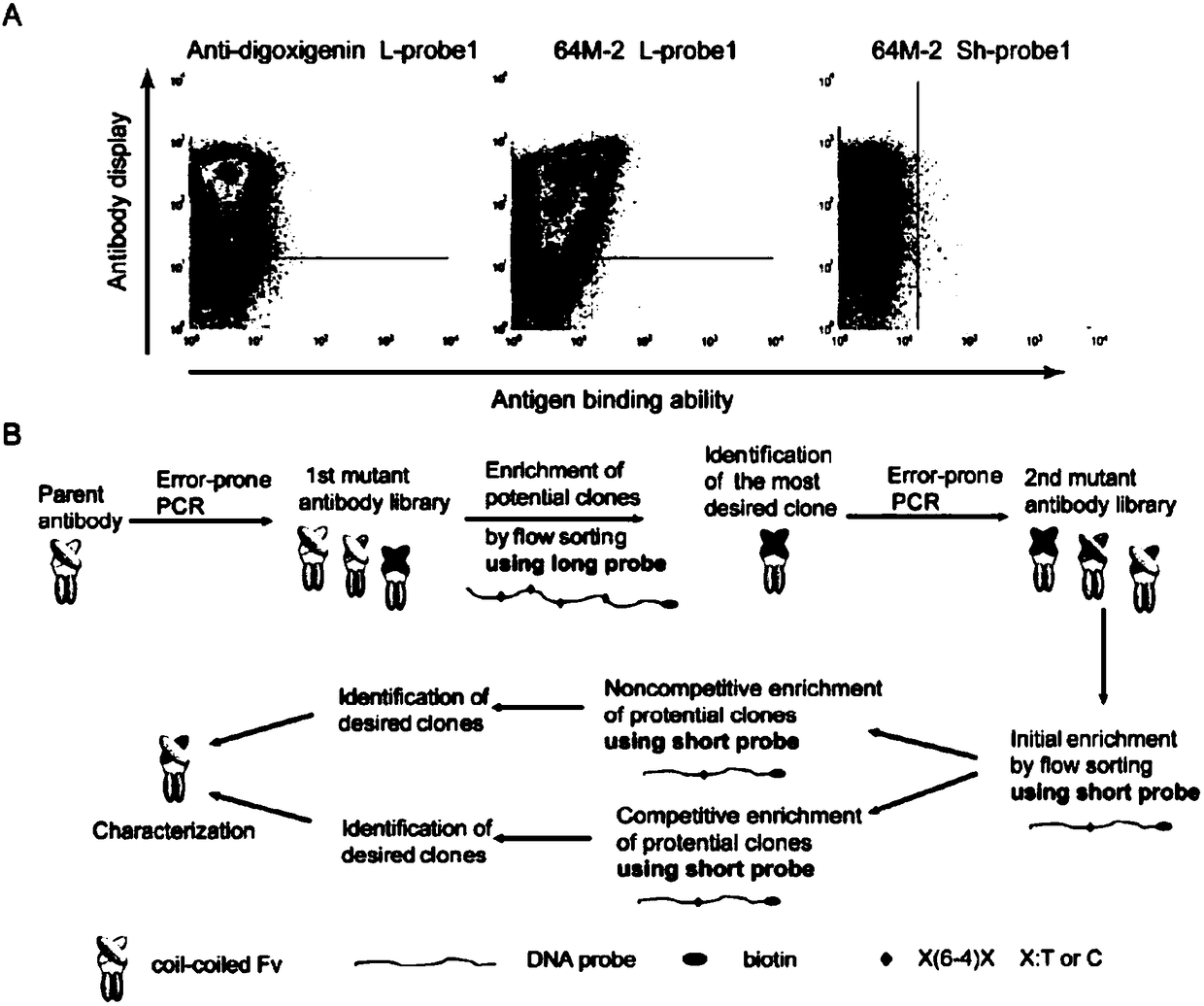
Antibody for identifying pyrimidine dimer generated by DNA irradiated by ultraviolet
A pyrimidine dimer, ultraviolet technology, applied in the field of medicine and biology, can solve problems such as inaccuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1, construction of related plasmids:
[0043] The variable region gene sequences (specific sequences) of the antibodies 64M-2 and 64M-5 against (6-4) photoproducts (6-4PPs) were obtained from the RCSB protein database (http: / / www.rcsb.org) and provided by Jin Synthesized by Sri Company as the target fragment. On the basis of the plasmid previously constructed in our laboratory, conventional cloning methods were used to replace the VH and VL regions in the plasmid with the heavy and light chain variable regions of the 64M-2 antibody by PCR or overlap PCR.
[0044] Among them, 64M-2VH was inserted into the sites of EcoRI and ClaI, 64M-2VL was inserted into the sites of EcoRI and KpnI, and finally two plasmids capable of displaying 64M-2ccFv on the inner membrane of E. coli were obtained. For detailed plasmid construction methods, please refer to the paper "Efficient Escherichia coli Antibody Display and Establishment of Artificial Evolution Platform" published...
Embodiment 2
[0047] Example 2, preparation of photoproduct (6-4PP) probe and purification of probe containing 6-4PP by HPLC:
[0048] 1. Probe preparation
[0049] In order to meet the requirements of flow sorting and detection, we prepared two kinds of probes containing 6-4PP: a long-chain probe (L-probe) containing 36 bases and a long-chain probe (L-probe) containing 10 bases Short chain probe (Sh-probe). The sequences of all probes can be found in Table 1. Because flow cytometry requires the detection of fluorescent signals, we conjugated biotin to the 3' end of all flow sorting and detection probes, which can be labeled with fluorescent molecularly labeled probes with streptavidin .
[0050] In Table 1, except that Sh-probe5 was purchased from TriLink BioTechnologies, all other probe sequences were synthesized by Shanghai Boshang Biotechnology Co., Ltd. The long-chain probe sequence L-probe1-8 was dissolved in double-distilled water to a final concentration of 50 μ M. Except for L-...
Embodiment 3
[0060] Example 3. Based on the 64M-2 antibody, artificial evolution of high-quality antibodies against photoproducts (6-4PP)
[0061] 1. Construction and evolution of 64M-2 antibody library
[0062] Select the V region gene sequence of the 64M-2 antibody as a template, first clone the VH and VL of 64M-2, and then use error-prone PCR to obtain an original antibody library with an average of 4-6 mutations in each gene . The primer pairs we used here are:
[0063] 5'-CGGACTACAAAGATGAATTCGAAGTTCAGCTGCAGCAGAG and
[0064] 3'-CCGCCGCCCTCGAGGTCGACTTTAATTTCCAGTTTGGTGCC.
[0065] We made some adjustments according to the manufacturer's instructions (Takara, Japan):
[0066] PCR buffer (50ul) contains:
[0067] 1ul low-fidelity polymerase (rTaq, Takara),
[0068] 40ng 64M-2 gene template,
[0069] Primer (10mM),
[0070] MnCl 2 (5mM), MgCl 2 (25mM), dNTP (2.5mM) and 20mM each of dCTP and dTTP.
[0071] The PCR program is:
[0072] (1) 94°C: 30s, (2) 55°C: 30s, (3) 72°C, 45s,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com