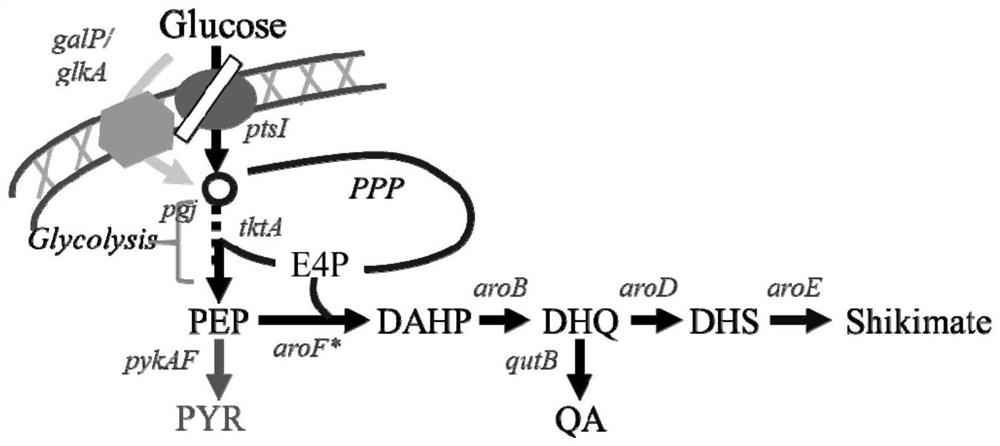
Recombinant Escherichia coli strain related to production of quinic acid and its construction method and application
A technology of recombinant strains and Escherichia coli is applied in the biological field to achieve the effects of reducing production costs, stabilizing heredity, and reducing the cost of culture media
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0147] The construction of embodiment 1 escherichia coli recombinant strain QA05
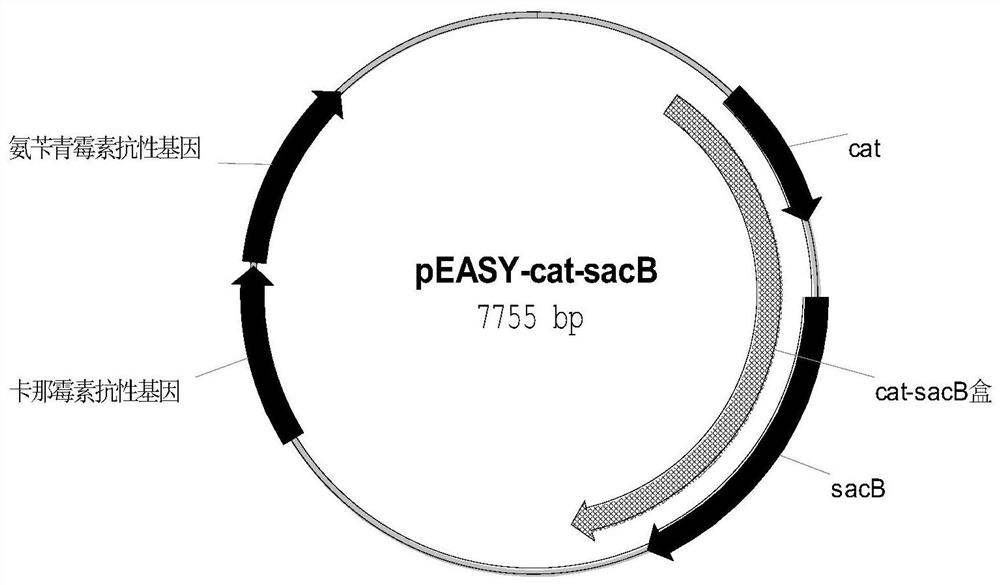
[0148] With the plasmid pEASY-cat-sacB (as shown in SEQ ID NO:2) containing chloramphenicol resistance gene cat and fructan sucrose transferase gene sacB figure 2 (shown) is a template, and primer 701aroD-cat-sacB-s / aroD-cat-sacB-a is used to amplify the fragment aroD1 of homologous recombination in the first step. The sequence of primer 701 is:
[0149] aroD-cat-sacB-s:ggtcatggggttcggtgcctgacaggctgaccgcgtgcaggtgacggaagatcacttc
[0150] aroD-cat-sacB-a:cccgcaccaatgacgagatcttttacagttacggttttcaatcaaagggaaaactgtcc
[0151] Amplification system: 5×TransStart TM FastPfu Buffer 10 μL, dNTPs (2.5mmol / L each dNTP) 4 μL, DNA template 1 μL (20-50ng), forward primer (10 μmol / L) 2 μL, reverse primer (10 μmol / L) 2 μL, 100% DMSO 1 μL, TransStart TM FastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL.
[0152] The amplification conditions are: pre-denaturation at 94°C for 5...
Embodiment 2
[0172] The construction of embodiment 2 escherichia coli recombinant strain QA07
[0173] With the plasmid pEASY-cat-sacB (as shown in SEQ ID NO:2) containing chloramphenicol resistance gene cat and fructan sucrose transferase gene sacB figure 2 (shown) is a template, and the primer 707acrB-cat-sacB-s / acrB-cat-sacB-a is used to amplify the fragment acrB1 of the first step of homologous recombination. The sequence of primer 707 is:
[0174] acrB-cat-sacB-s:ctgtcagaattgggtatattggggcaggttgtcgtgaaggaattccctagtgacggaagatcacttc
[0175] acrB-cat-sacB-a:agttttccctggtgttggcgcagtattcgcgcaccccggtctagccggggatcaaagggaaaactgtc
[0176] Amplification system: 5×TransStart TM FastPfu Buffer 10 μL, dNTPs (2.5mmol / L each dNTP) 4 μL, DNA template 1 μL (20-50ng), forward primer (10 μmol / L) 2 μL, reverse primer (10 μmol / L) 2 μL, 100% DMSO 1 μL, TransStart TM FastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL.
[0177] The amplification conditions are: pre-dena...
Embodiment 3
[0198] Example 3 Construction of Escherichia coli recombinant strain QA10
[0199] With the plasmid pEASY-cat-sacB (as shown in SEQ ID NO:2) containing chloramphenicol resistance gene cat and fructan sucrose transferase gene sacB figure 2 (shown) is a template, and primer 1001 2-ackA-pta-cat-sacB-s / 2-ackA-pta-cat-sacB-a is used to amplify the fragment ackA-pta1 of the first step of homologous recombination. The sequence of primer 1001 is:
[0200] 2-ackA-pta-cat-sacB-s:ctgacgtttttttagccacgtatcaattataggtacttcctcctggtgtccctgttgatacc
[0201] 2-ackA-pta-cat-sacB-a:ttcagatatccgcagcgcaaagctgcggatgatgacgagaatagatacatcagagcttttacgag
[0202] Amplification system: 5×TransStart TM FastPfu Buffer 10 μL, dNTPs (2.5mmol / L each dNTP) 4 μL, DNA template 1 μL (20-50ng), forward primer (10 μmol / L) 2 μL, reverse primer (10 μmol / L) 2 μL, 100% DMSO 1 μL, TransStart TM FastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL.
[0203] The amplification conditions a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com