Application of specific methylation sites as breast cancer molecular subtype diagnosing markers
A molecular typing and methylation technology, applied in the determination/examination of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve problems such as non-minimally invasive or non-invasive breast cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
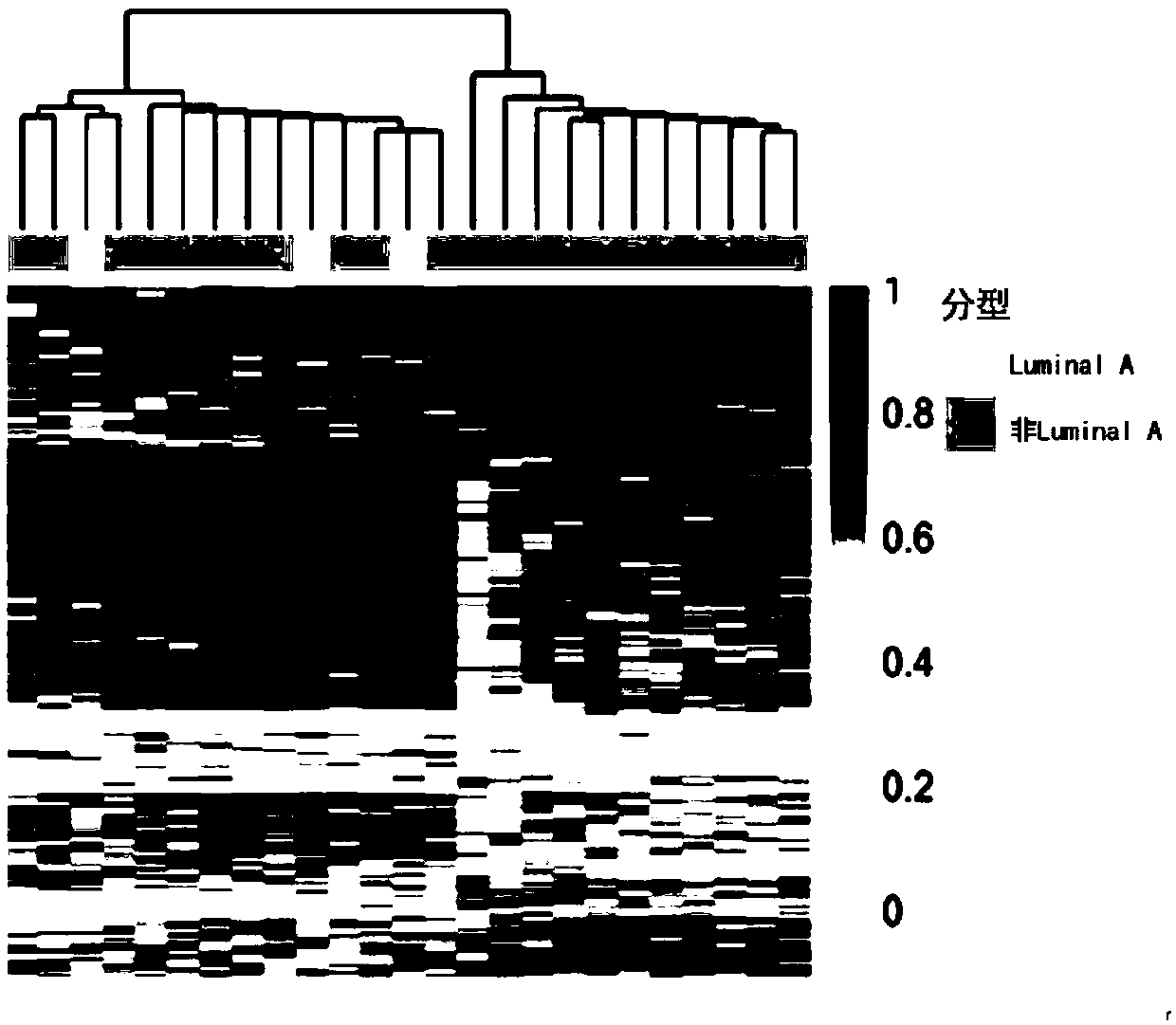
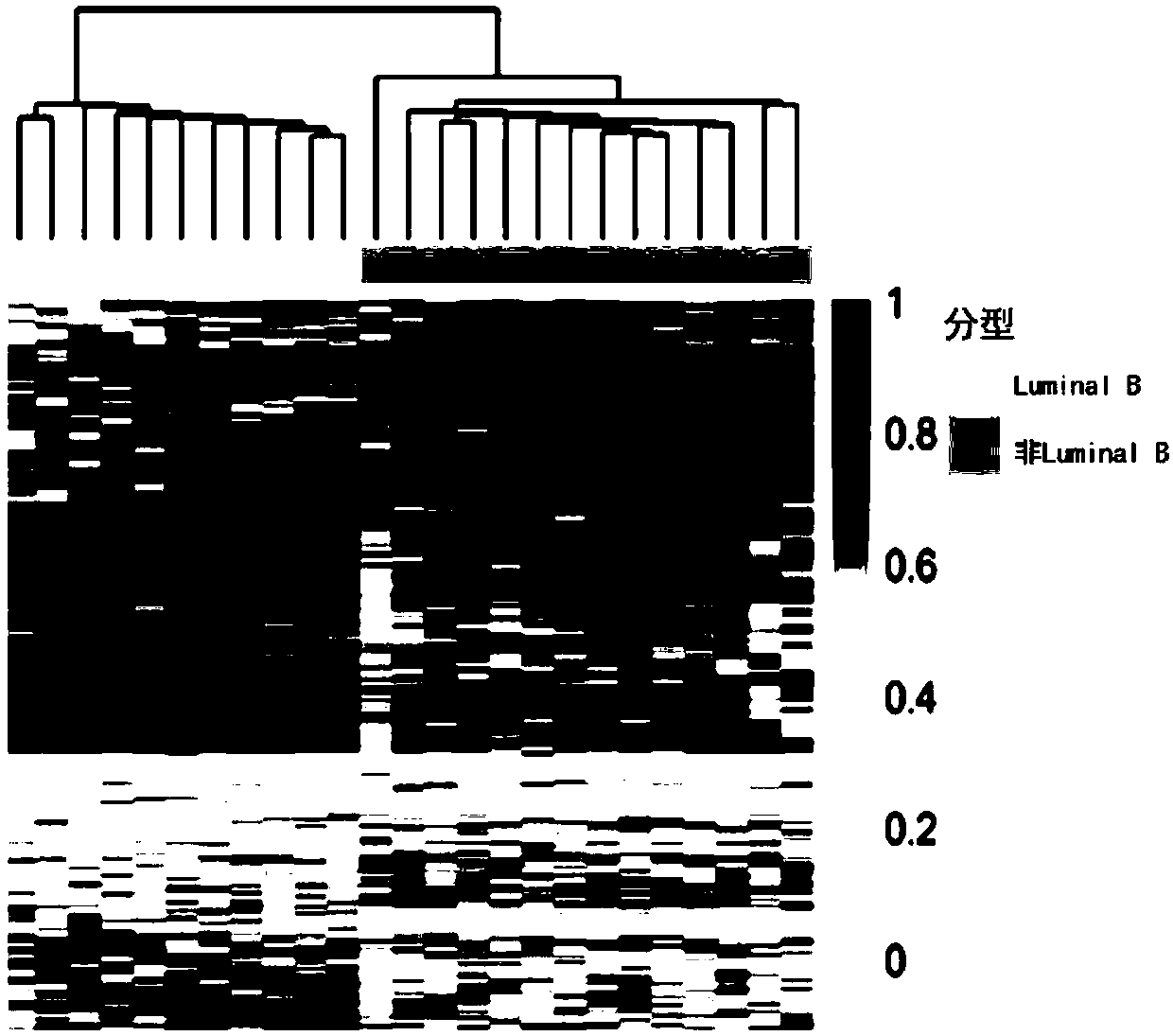
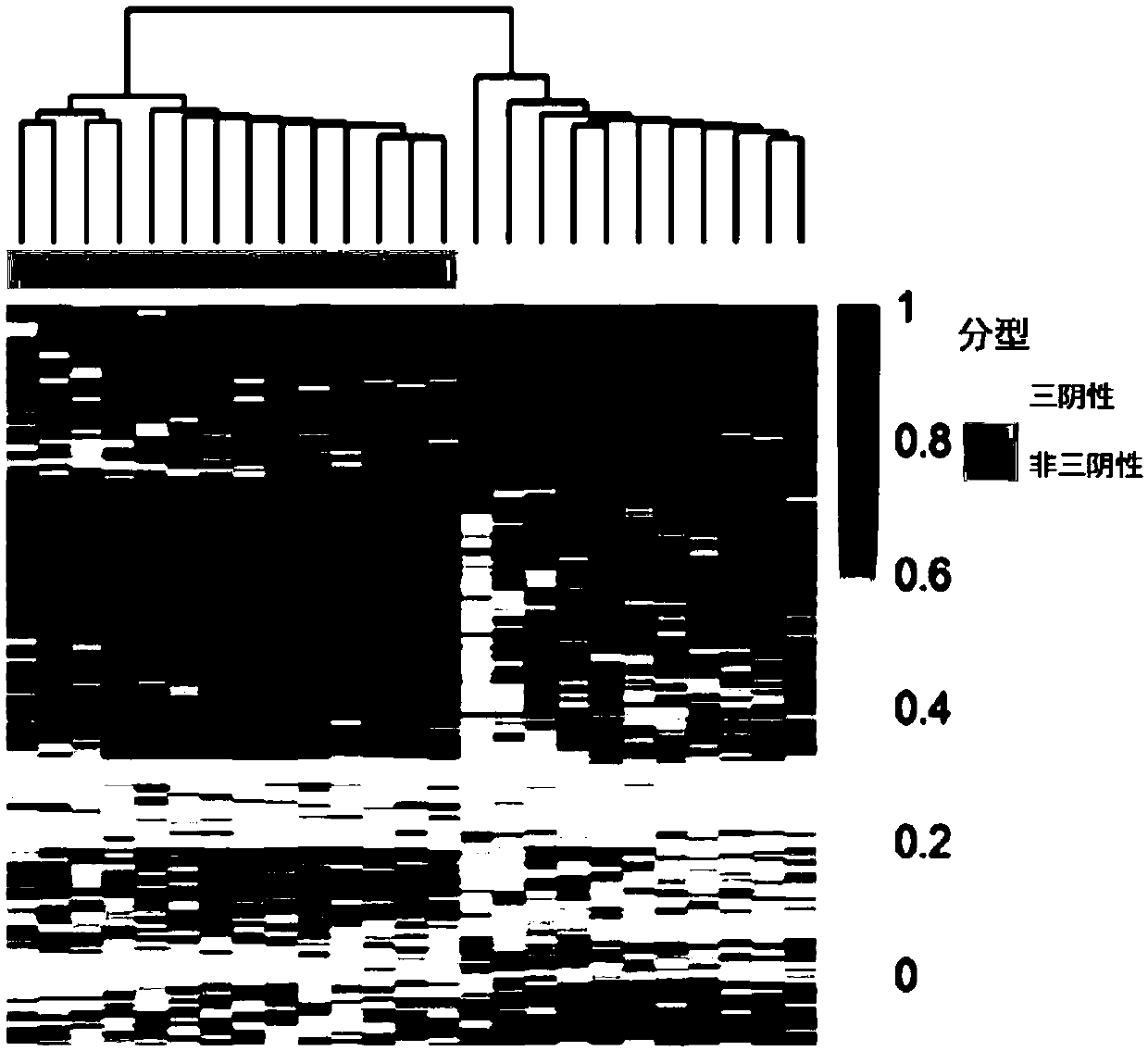
[0078] According to a typical embodiment of the present invention, the device further includes an evaluation module, which is used to compare the detection value of the subject with the detection value of breast cancer patients with different molecular types, when selected from chr10: 39097581, chr10: 88386316, chr11: 105772483,chr11:113528661,chr1:163455492,chr1:164069493,chr1:173149222,chr1: 201310696,chr12:29221374,chr1:233270329,chr1:242355017,chr14:60666657,chr1: 59426963,chr16:46420219,chr18: 61618871,chr21:20247091,chr21:23757716,chr2: 36975610,chr2:87314115,chr2:89834041,chr2:89837315,chr2:89858946,chr3:115818550, chr3:144975142,chr4:164139191,chr4:189484234,chr4:3579745, chr4:47297495,chr4: 71802931,chr5:42079715,chr6:100477543,chr6:153594933,chr6:40877403,chr6:75723365, chr7:103135406,chr7:148667863,chr7:43817571,chr8:27578331,chr8:46896342和chr8: The methylation changes of one or more sites in 75989278 are in the same trend compared with the detected values of pati...
Embodiment 1
[0098] Collection and identification of blood samples from breast cancer patients
[0099] Blood samples from patients with different molecular types were collected and collected through verification by experienced physicians (the samples were provided by Tianjin Cancer Hospital).
Embodiment 2
[0101] Extraction and library sequencing of cfDNA in breast cancer patients
[0102] Blood samples from patients with various breast cancer molecular types were collected, and cfDNA was extracted from plasma. Utilize the QIAamp Circulating Nucleic Acid Kit.
[0103] The specific operation steps are as follows: 1) Use the density gradient centrifugation method to centrifuge at a speed of 1900g to separate red blood cells, white blood cells and plasma. The clear top layer is plasma, take out the plasma, and then use a speed of 14000rpm to perform high-speed centrifugation again. The upper layer is plasma; 2) Add the extracted plasma from 1) to a 50ml centrifuge tube that has been added with proteinase K, and then add the lysate ACL (which already contains CarrierRNA) to obtain the first mixed solution; 3) Add the last obtained second mixture in 2) 1. Mix the mixed solution and put it in a water bath at 60°C for 30 minutes; 4) Take out the first mixed solution from the water bat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com