Repositioning drug discovery method based on integration of a plurality of transcriptome data sets and drug target information
A discovery method and transcriptome technology, applied in the field of repositioning drug discovery, can solve problems such as gaps and costs that are difficult to cover diseases and drugs, and achieve the effects of low cost, clear principle and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
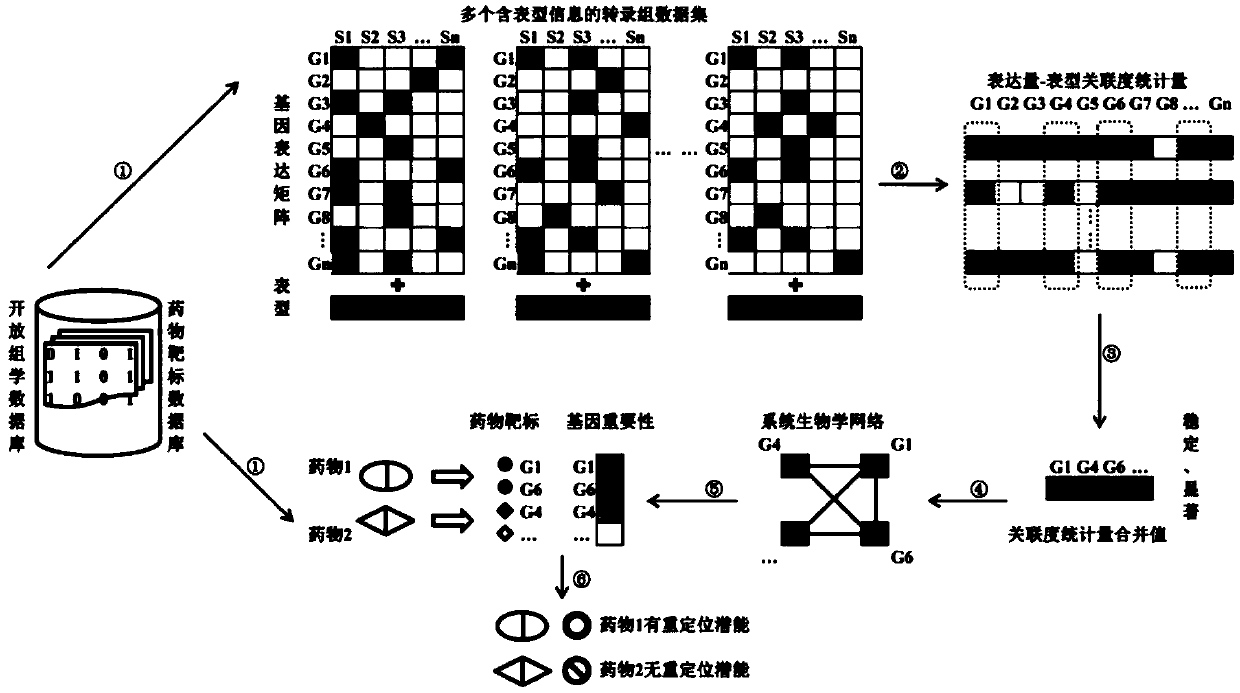
[0045] Example one, as figure 1 and figure 2 As shown, a drug discovery method based on the integration of multiple transcriptome datasets and drug target information, comprising the following steps:
[0046] Step 1: For any disease, obtain multiple transcriptome datasets containing phenotypic information from open omics databases;
[0047] Among them, the open omics databases are Gene Expression Omnibus (GEO, https: / / www.ncbi.nlm.nih.gov / geo / ), ArrayExpress (https: / / www.ebi.ac.uk / arrayexpress / ) and Genomic One or more of the Data Commons (GDC, https: / / portal.gdc.cancer.gov / ). The sources of these databases are authoritative and reliable, and the information and data are open to the public and can be obtained free of charge. The transcriptome data in the open omics database involves the transcriptome information of thousands of diseases, including a large number of human tissue transcriptome data and a large number of transcriptome data sets containing phenotypic informati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com