Expression vector of fusion protein for enhancing target RNA loading into exosome and establishment method and application thereof
An expression vector and fusion protein technology, applied in the field of molecular biology, can solve the problem of lack of effective means, and achieve the effect of reducing undesired RNA entry into exosomes, efficient loading, and reducing loading
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Construction of RNA-binding protein exosomes
[0048] The specific steps to build are as follows:
[0049] (1) Trizol extracts total RNA from mouse fibroblasts;
[0050] (2) Using Promega M-MLV reverse transcriptase to reverse transcribe RNA to obtain mouse fibroblast cDNA;
[0051] The reaction system is shown in Table 1:
[0052] Table 1 reaction system
[0053] RNA template
2μg
5×RT buffer
4μl
dNTP Mixture (10mM)
2μl
0.1M DTT
2μl
M-MLV reverse transcriptase
1μl
overall system
20μl
[0054] Mix the above reagents evenly, centrifuge at low speed for a short time, place in a 37°C incubator for 1 hour, then inactivate at 80°C for 5 minutes, and store at -20°C.
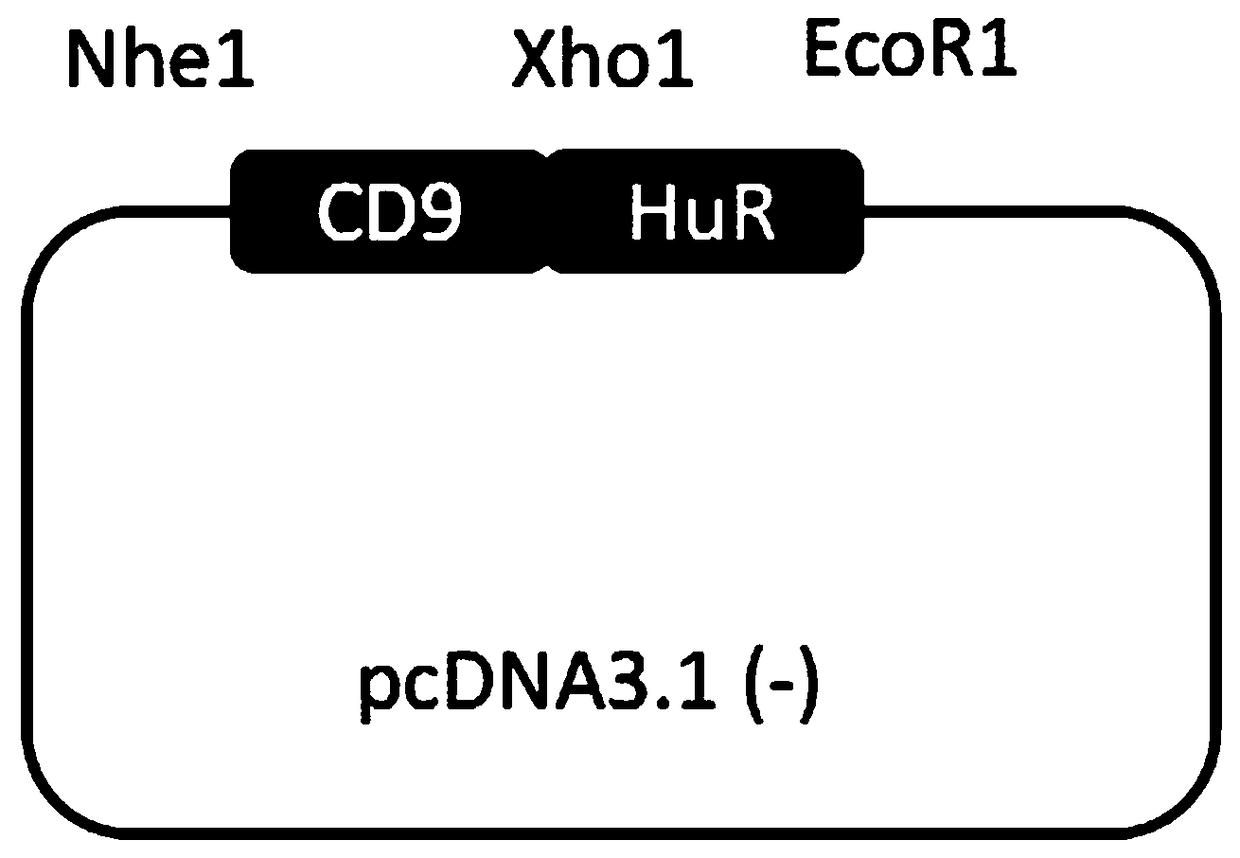
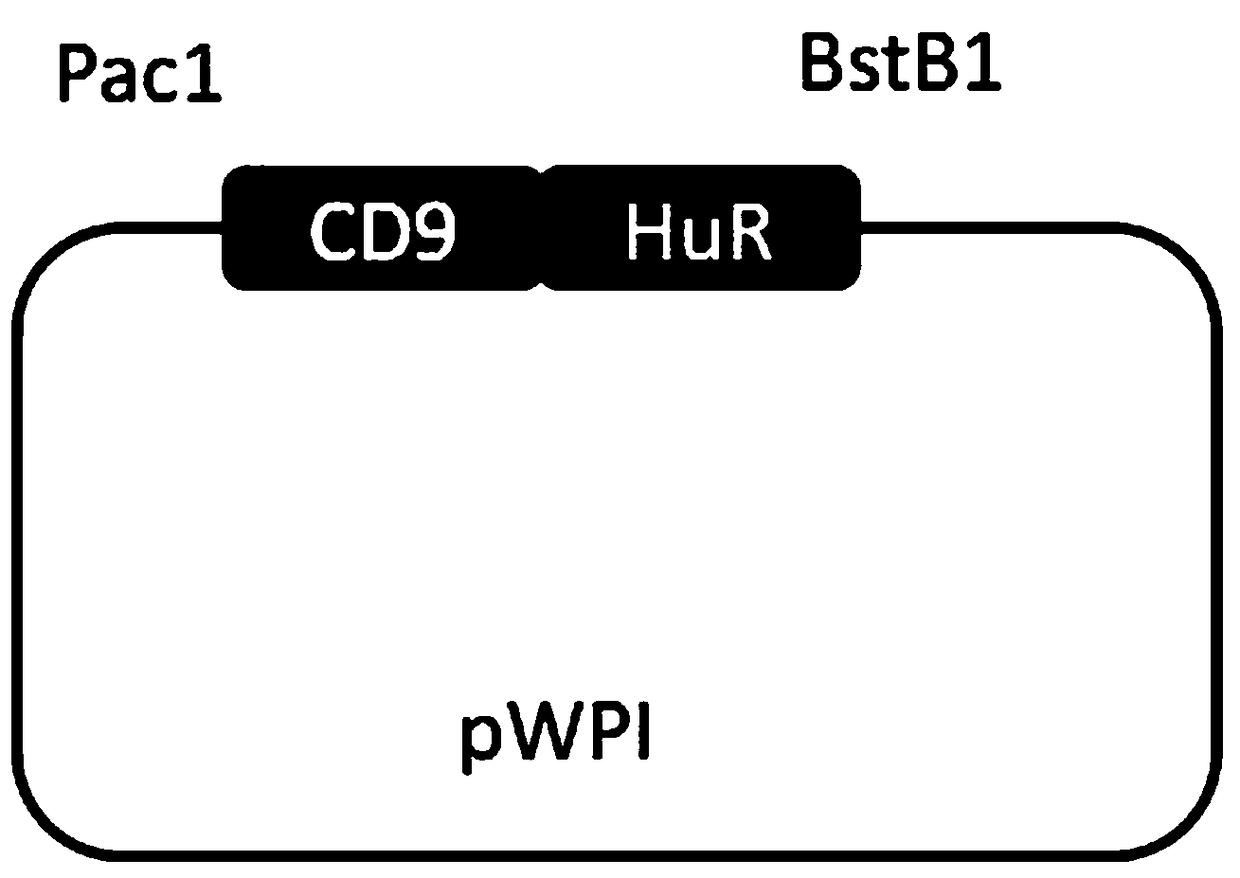
[0055] (3) Using the reverse transcription cDNA as a template, clone the CDS region of the RNA binding protein HuR and the CDS region of the membrane molecule CD9;
[0056] The primers and PCR reaction system are shown in Tab...
Embodiment 2
[0074] Example 2: Verification test of physicochemical properties of modified exosomes
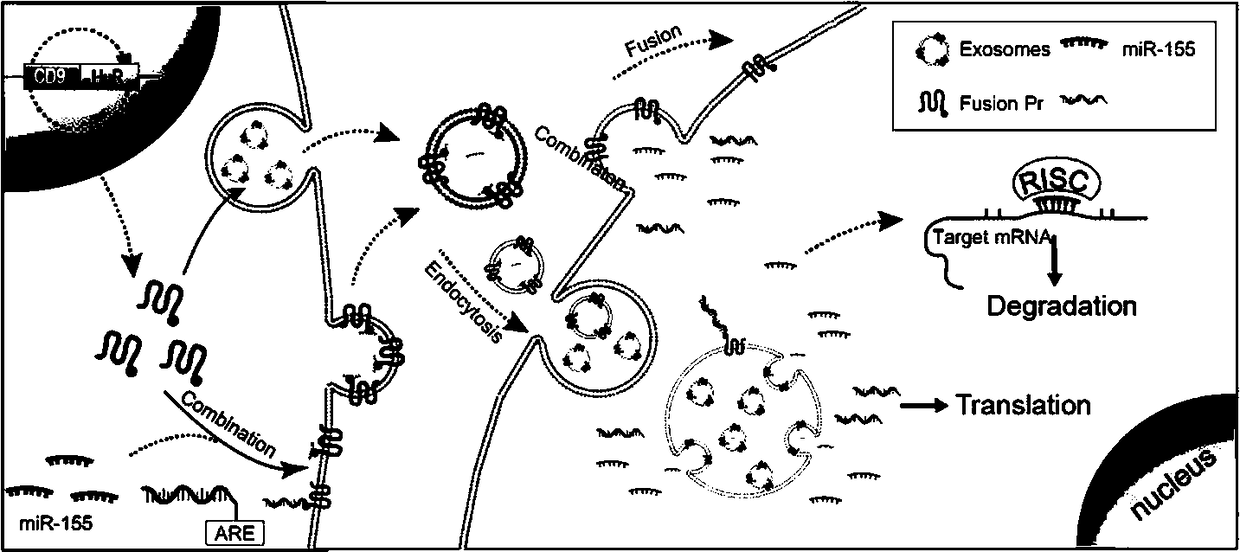
[0075] like Figure 4 As shown, the viral packaging cell HEK293T was infected with the virus expressing the fusion protein CD9-HuR constructed in Example 1 to obtain virus particles expressing the fusion protein. Bone marrow mesenchymal cells MSC were infected with the virus, and then cultured under serum-free conditions, exosomes were collected between 48 and 72 hours, and exosomes were extracted through the exosome isolation and extraction kit ExoQuick. Identify the number, size, and fusion expression efficiency of exosomes.
[0076] like Figure 5 As shown, the collected exosomes were diluted with PBS at 1:1000, and then the number and size of exosomes were identified by Nanosight, and it was found that the size of exosomes was within the range of 40-200nm.
[0077] like Image 6 As shown, the shape of the exosomes was further clarified by electron microscopy, and the modified exoso...
Embodiment 4
[0079] Example 4: Nucleic acid loading test of modified exosomes
[0080] Infect the exosome packaging cells with the constructed virus expressing the fusion protein CD9-HuR, then transfect the exosome packaging cells with miR155 or dCas9-ARE to be loaded, and then culture them under serum-free conditions for 48 to 72 hours. Exosomes were collected in between, and exosomes were extracted by exosome separation and extraction method.
[0081] (1) Loading and delivery of miR155
[0082] miR155 was directly synthesized.
[0083] The constructed virus expressing the fusion protein CD9-HuR was used to infect the exosome packaging cells, and then the exosome packaging cells were simultaneously transfected with miR155 to be loaded, then cultured under serum-free conditions, and the exosome packaging cells were collected between 48 and 72 hours. Exosomes are extracted by exosome isolation and extraction methods.
[0084] The loading, delivery and intervention efficiency of exosomes ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com