Nucleic acid detection method based on prokaryotic argonaute protein and its application
一种核酸、靶标核酸的技术,应用在生物领域
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
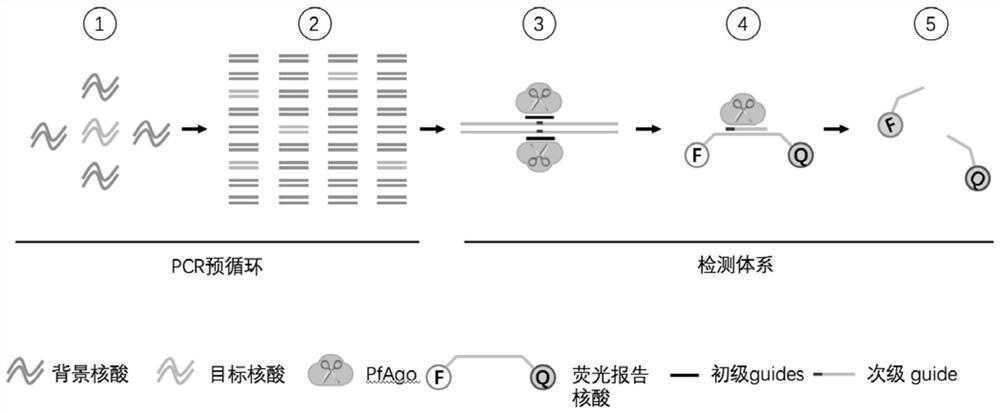
[0158] Preparation of detection reagent and detection method
[0159] In this embodiment, a kit for the nucleic acid detection method based on the gene editing enzyme Pyrococcus furiosus Argonaute (PfAgo) of the present invention and its usage method are provided.
[0160] 1.1 Detection reagents and kits
[0161] In this embodiment, taking the detection of the E545K mutation of the PIK3CA gene as an example, the corresponding specific target nucleic acid sequence is 5'-CTGTGACTCCATAGAAAAATCTTTCTCCTGCTCAGTGATTTCAGAGAGAGGATCTCGTGTAGAAATTGCTTTGAGCTGTTCTTTGTCATTTTCCCCT-3', SEQ ID No.: 1.
[0162] Based on the method of the present invention, the corresponding detection reagents include the following:
[0163] (1), amplification primer F-primer and R-primer, specific sequence is as follows:
[0164] F-primer: 5'-CTGTGACTCCATAGAAAATCTTTCTCC-3' (SEQ ID No.2)
[0165] R-primer: 5'-AGGGAAAATGACAAAGAACAGCTC-3' (SEQ ID No.3)
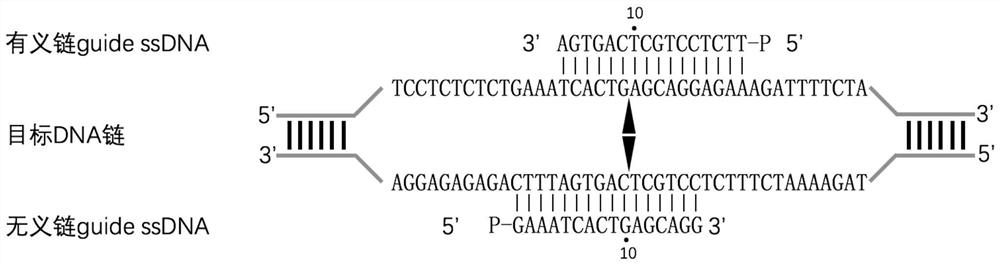
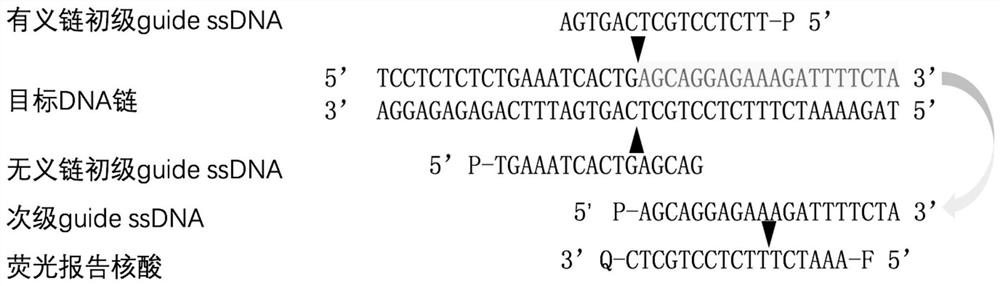
[0166] (2), specific guide ssDNAs pair, including sense s...
Embodiment 2
[0182] Detection of different concentrations of nucleic acids to be detected
[0183] The specific target nucleic acid (SEQ ID NO.: 1) was diluted according to the principle of 10-fold dilution method, and diluted into standard mother solutions of 200pM, 20pM, 2pM, 200fM, 20fM, 2fM and OfM respectively. Nucleic acid standard mother solutions of different concentrations were added to the reaction system described in Example 1, and the sample addition reaction was carried out step by step, and the fluorescent signal value at the wavelength of the corresponding fluorophore was detected by fluorescent quantitative PCR.
[0184] The result is as Figures 4A-4H shown. in, Figure 4A , 4B The concentration of the target nucleic acid in , 4C, 4D, 4E, 4F, 4G is 0fM, 2fM, 20fM, 200fM, 2pM, 20pM, 200pM respectively, Figure 4H It is a non-target nucleic acid (20ng is added to the system).
[0185] Non-target nucleic acid is the total DNA extracted from normal human serum. The resul...
Embodiment 3
[0188] Specificity tests and multiplexing
[0189] Prepare different types of target nucleic acid solutions with a concentration of 200pM, which are two types of HCV virus, JFH-1 2a and CON1-1b.
[0190] It should be noted that the difference between the two types is only that there are two consecutive differences in the sequence.
[0191] Configure a multiplex mixed detection system: add 2 pairs of specific amplification primers to the detection amplification system at the same time, and add the target detection nucleic acid to the detection amplification system in the following 4 groups: (1) blank control; (2) JFH- 12a; (3) CON1-1b; (4) JFH-1 2a and CON1-1b. (The specificity of the fluorescent reporter nucleic acid is verified respectively in Figure 5, and they do not affect each other in a reaction system)
[0192] After the amplification reaction, the PfAgo enzyme, MnCl 2 , 2 groups of specific ssDNAs, 2 pairs of fluorescent reporter nucleic acids with different fluores...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com