Scaffolding method based on long readings and contig classification
A reading and partial technology, applied in the field of sequence assembly in bioinformatics, can solve problems such as high sequencing error rate, noisy comparison information, and affecting the accuracy of scaffolding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
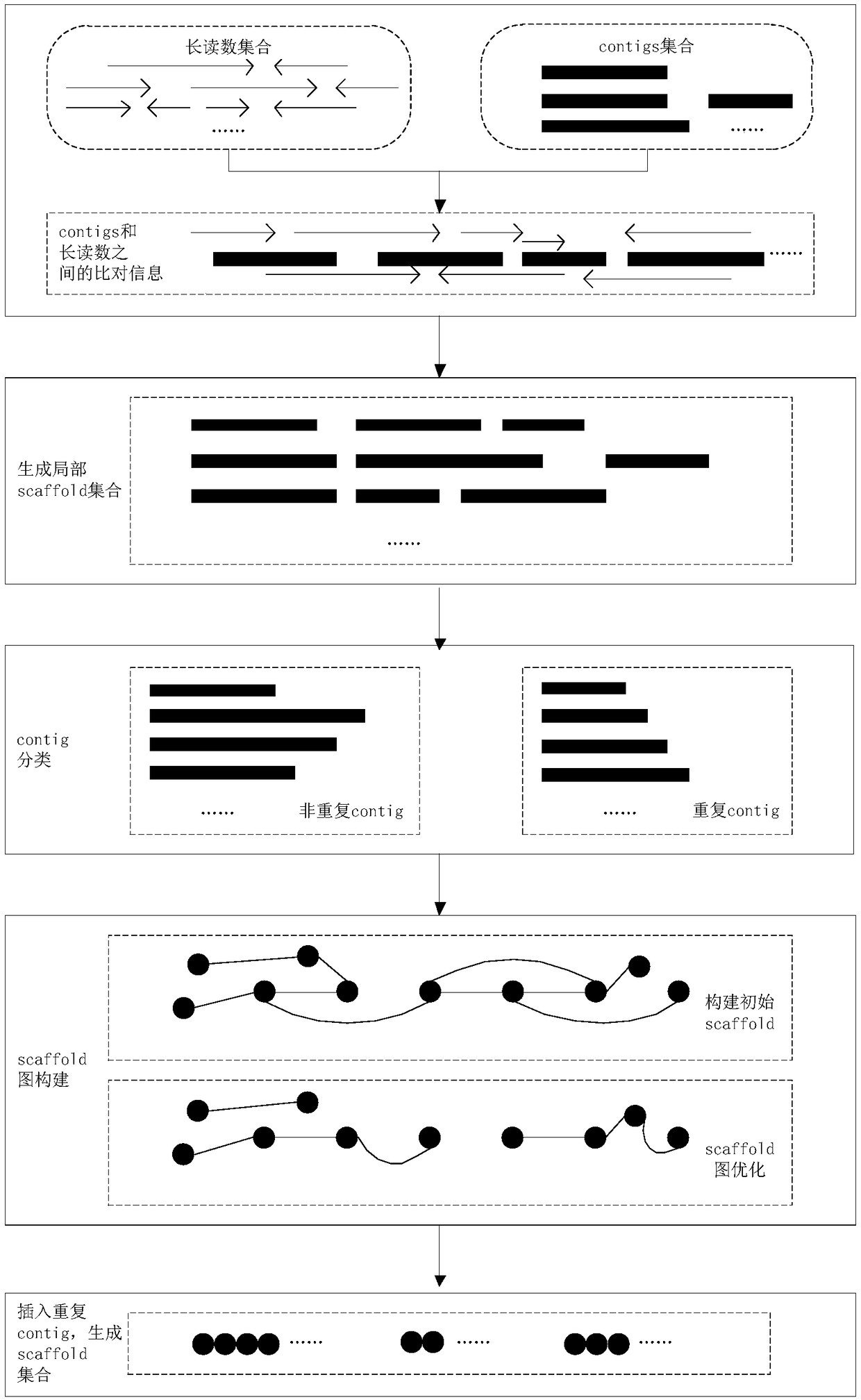
[0061] like figure 1 As shown, the specific implementation process of the present invention is as follows:
[0062] 1. Generate a local scaffold set
[0063] 1.1 This method takes contig files and long read files as input data. First, use the alignment tool BWA to align the long reads to the contig to obtain the alignment result. where only lengths greater than L are considered r of long reads and lengths greater than L c contig, L r =500, L c = 3000.
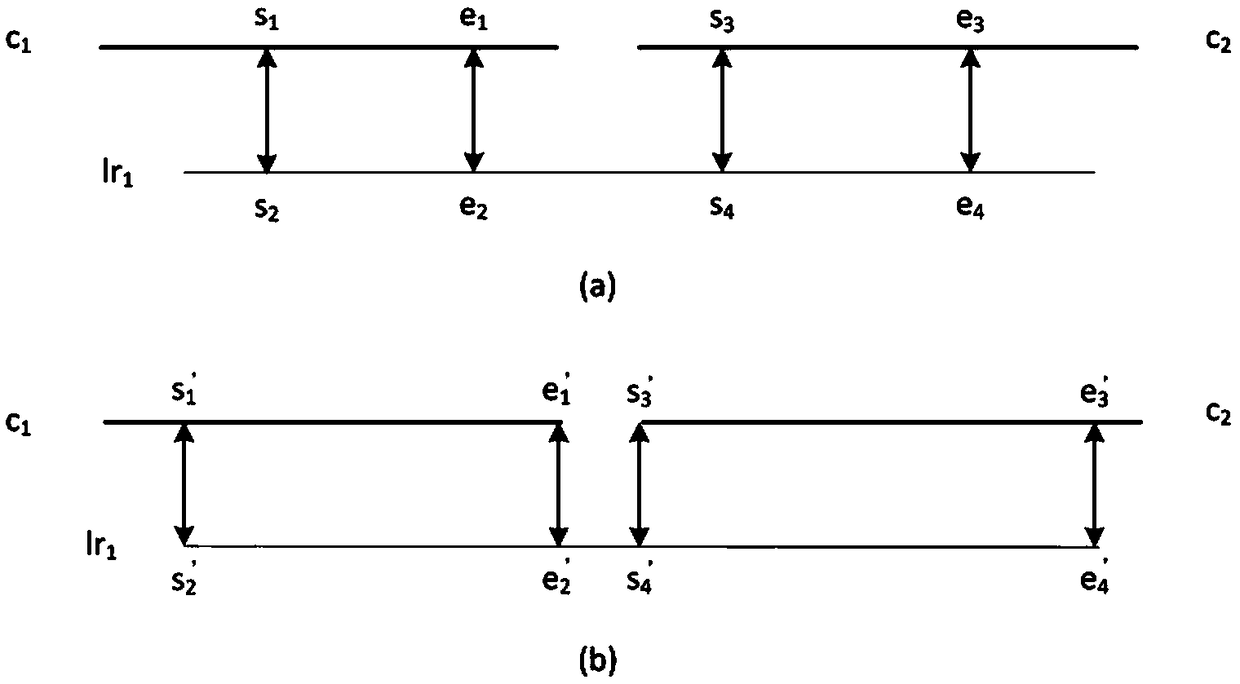
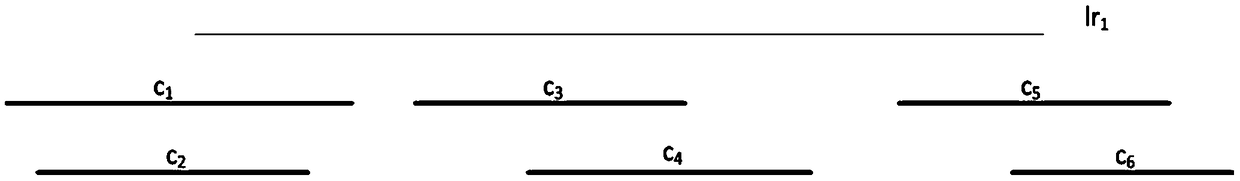
[0064] 1.2 If a long read and a contig can be aligned, the position and alignment direction of the alignment interval can be obtained. Assuming the jth long read (lr j ) and the ith contig (c i ) can be compared, it means lr j The previous interval can be compared to c i an interval above. This method uses SPR (c i ,lr j ) in lr j The starting position of the above alignment interval, EPR(c i ,lr j ) in lr j The end position of the upper alignment interval, SPC(c i ,lr j ) represents the starting position of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com