Quick detection method for SNP loci of peripheral blood leucocytes
A detection method and white blood cell technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., to achieve good application prospects, simple operation process, and short operation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
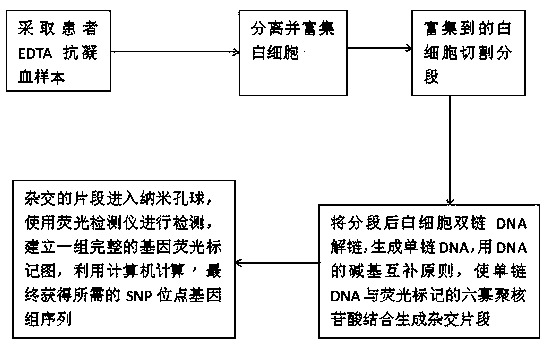
[0015] figure 1 As shown in , a rapid detection method for SNP sites in peripheral blood leukocytes, using ammonium chloride solution, nanoporous spheres, and hexaoligonucleotides as the supplies used for detection, ammonium chloride solution, nanoporous spheres, hexaoligonucleotides The composition ratios of the nucleotide components are: 0.74% ammonium chloride solution, 0.3% nanoporous balls, and 10UM hexaoligonucleotide.
[0016] figure 1 As shown in , a rapid detection method for SNP sites in peripheral blood leukocytes, the detection of SNP sites in peripheral blood leukocytes is completed through the following steps. The first step is to take the patient's EDTA anticoagulated blood sample; The amount is 2ml. The second step is to separate and enrich white blood cells; the process of separating and enriching white blood cells is as follows, put the ammonium chloride solution and 2 ml of the patient's DTA anticoagulant blood sample into the test tube and mix well, after...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com