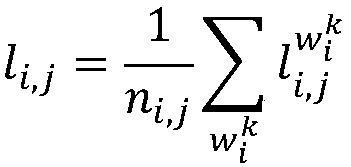Self-avoiding random walk-based disease-associated miRNA prediction method and system
A random walk and prediction method technology, applied in the field of systems biology, can solve the problems of expensive and time-consuming biological experimental methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035]In the present invention, the degree of association between the query disease and the candidate miRNA is defined as the product of the reciprocal of the average step length from the query disease to the candidate miRNA and the transition probability from the query disease to the candidate miRNA, and its expression is as follows:
[0036]
[0037] Among them, p i,j is the disease to be queried d i to miRNA m j Transition probability, l i,j is the disease to be queried d i to miRNA m j Average step size.
[0038] The whole process of a disease-associated miRNA prediction method based on self-avoiding random walk is as follows: figure 1 shown. First input a set of miRNA-disease association information, this method includes the following sub-processes:
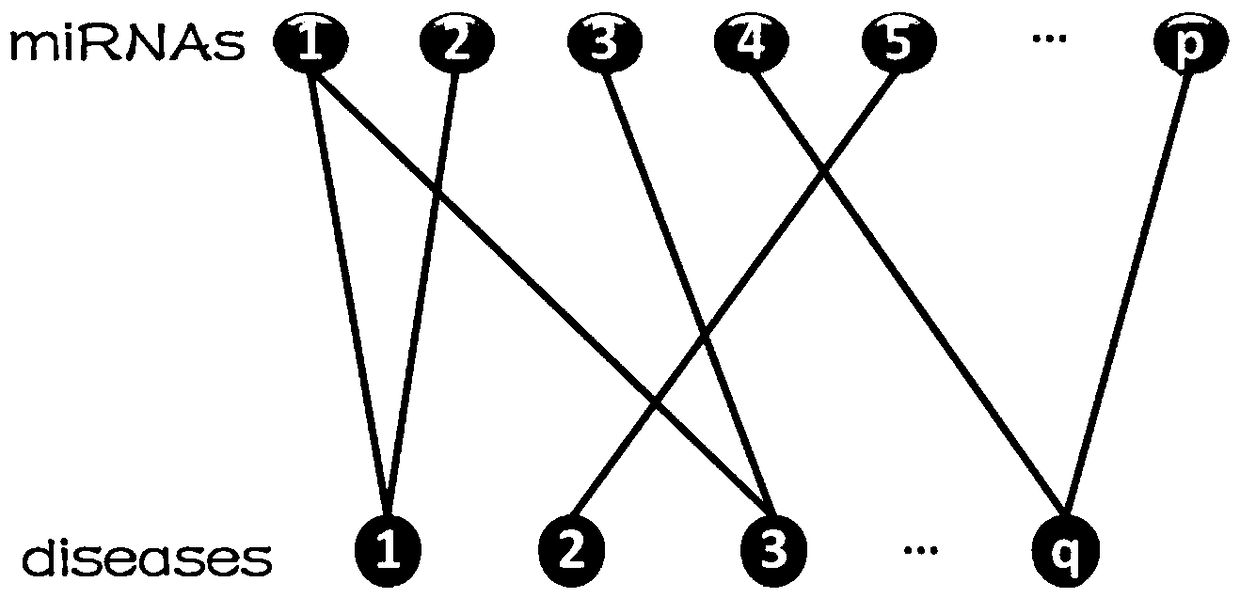
[0039] 1) Establish miRNA-disease bipartite graph: input a set of miRNA-disease association information, and establish miRNA-disease bipartite graph G= (such as figure 2 shown);
[0040] Among them, miRNA-diseas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com