Method for knocking out plutella xylostella APN1 gene with CRISPR/Cas9 gene editing system and application of method
A gene knockout and Plutella xylostella technology, applied in the field of genetic engineering, can solve the problems of cumbersome identification process and insufficient verification methods, and achieve the effect of simple operation and high gene editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
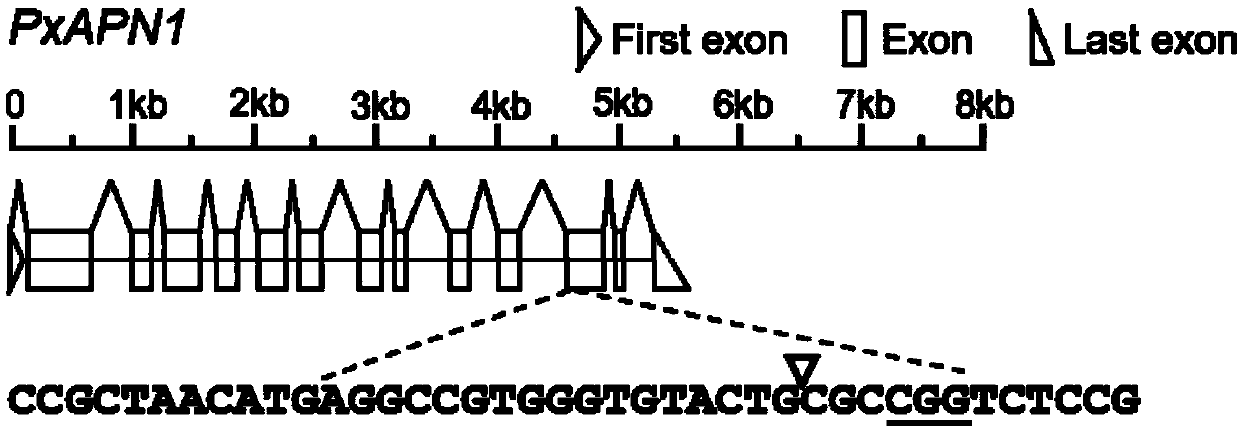
[0020] Design and synthesis of sgRNA target sequence of Plutella xylostella APN1 gene
[0021] 1. Use the Cas-Designer software (http: / / www.rgenome.net / cas-designer / ) to design the sgRNA target sequence (the nucleotide sequence) in the specific region (the 12th exon) of the Plutella xylostella APN1 gene Shown in SEQ ID NO. 1), such as figure 2 shown, and search the diamondback moth DBM-DB genome database (http: / / 59.79.254.1 / DBM / index.php) and CRISPR off-target effect detection Cas-OFFinder software (http: / / www.rgenome.net / cas-offinder / ), to detect potential off-target sites.
[0022] 2. Add the T7 promoter sequence before the sgRNA core site AGGCCGTGGGTGTACTGCGC, and add the sequence complementary to crRNA / tracrRNA after the sgRNA core site to form the complete 5′-end DNA fragment of the sgRNA.
[0023] 3. The 80-bp crRNA / tracrRNA sequence and the sgRNA 5' end DNA fragment in step 2 undergo PCR denaturation, annealing, and extension to synthesize a complete sgRNA deoxynu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com