Method for rapidly detecting 3'-5' exo-activity of DNA polymerase or mismatching and kit thereof
A polymerase and kit technology, applied in the field of molecular biology, can solve the problems of inapplicability of sequencing enzyme exo-cutting activity detection and screening, inapplicability of sequencing enzyme detection and screening, inability to detect DNA polymerase exo-cutting activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
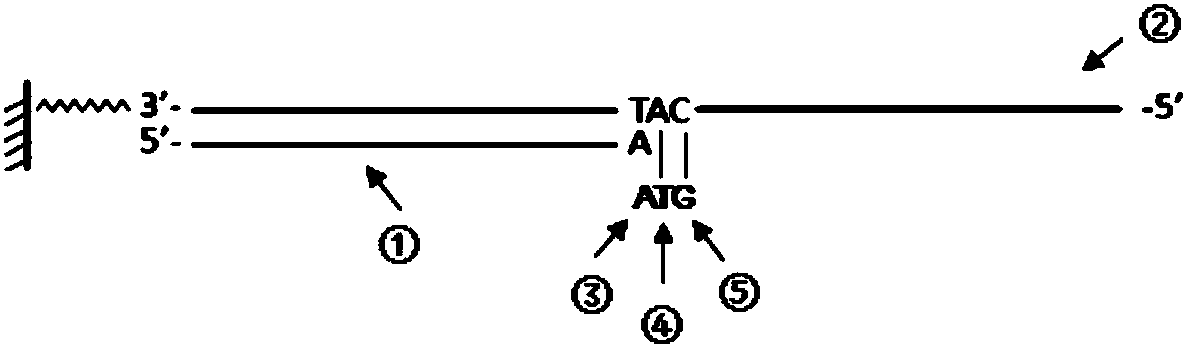
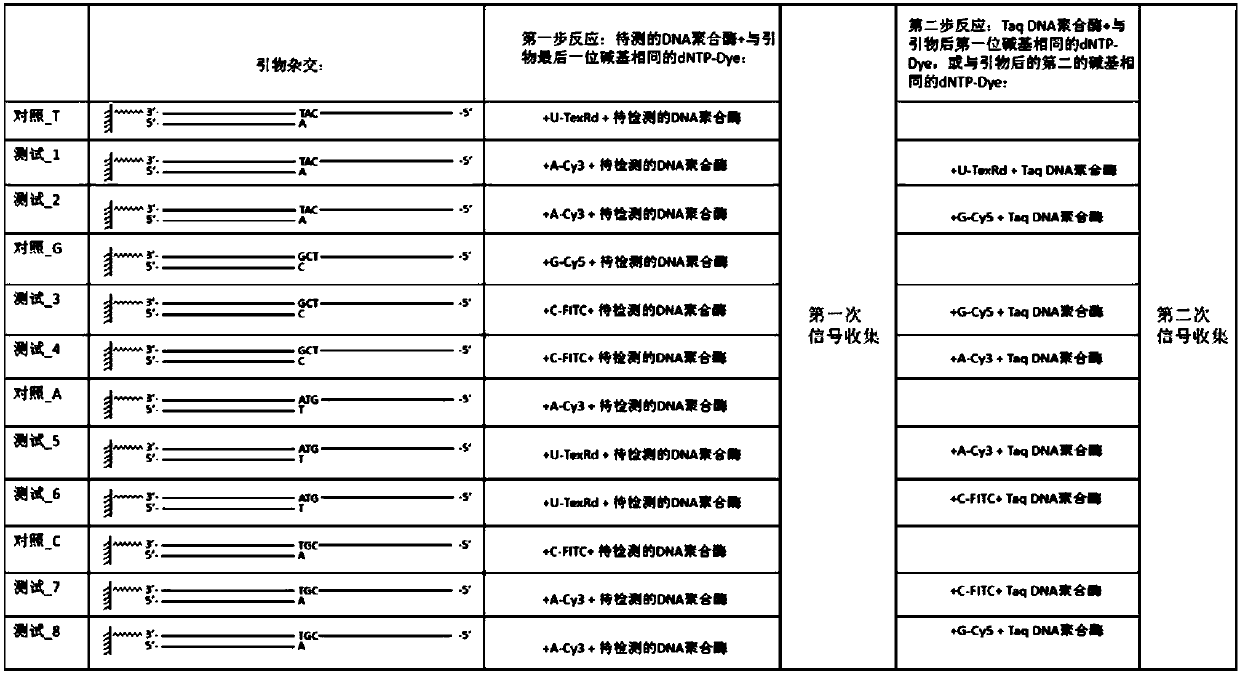
[0040] The DNA sequence (template) immobilized on the solid phase can be immobilized on the chip or magnetic beads, and this embodiment is carried out on the nucleic acid sequence (DNB, DNA nanoball) immobilized on the chip.
[0041] 1. Equipment:
[0042] Bgiseq1000 sequencer (BGI), sequencing chip (BGI), VWR heating plate, PCR machine, PCR eight-tube.
[0043] 2. Reagents:
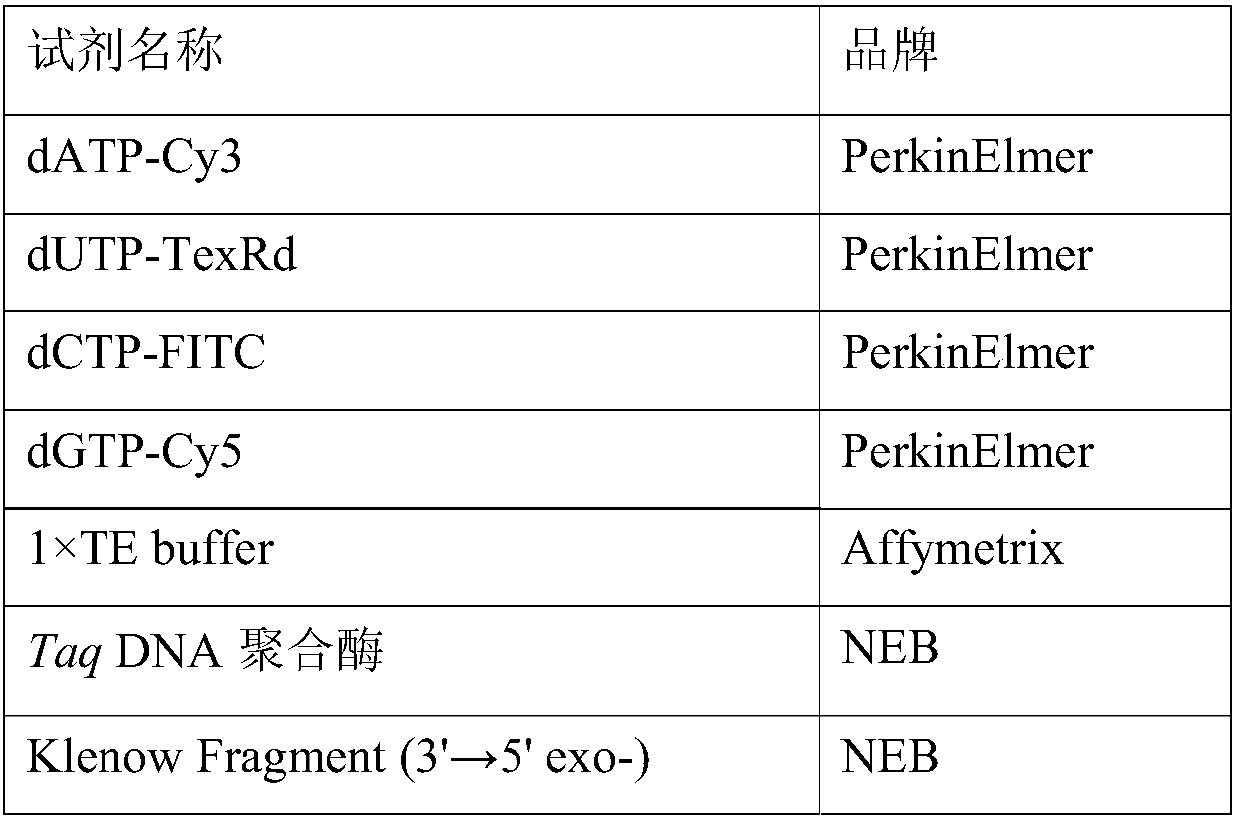
[0044] The reagents used in this example are shown in Table 1 below.
[0045] Table 1 Reagents
[0046]
[0047]
[0048] The fixed sequence on the DNB used in this embodiment is:
[0049] AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTGCAT (SEQ ID NO: 1).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com