Detection method and kit for detecting 1p/19q combination loss of heterozygosity of chromosome
A technology for lack of heterozygosity and detection methods, applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of unreliable analysis of markers, etc., achieve fast turnaround time, high sensitivity and specificity, and easy operation easy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
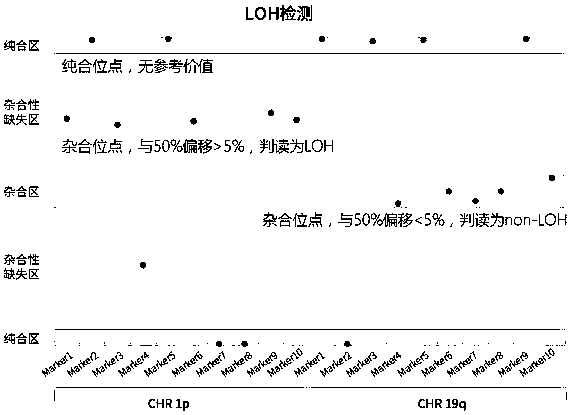
[0033] Example 1 High-throughput next-generation sequencing detection method of Chr1p / 19q co-LOH based on multiple amplicons
[0034] 1. Site selection
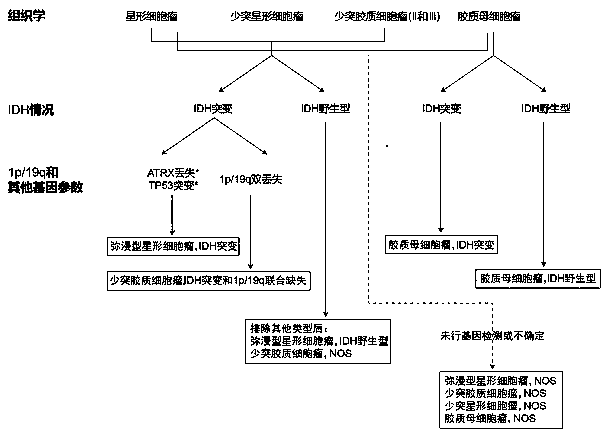
[0035] Select 10 SNPs on the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q), among which, 5 SNPs on 1p are located in the 1p36.3 segment, and 5 SNPs on 19q are located in the 19q13.3 segment Segment, these two regions are used to determine whether loss of heterozygosity occurs in the common deletion segment, combined with other SNPs, it can be used to determine whether loss of heterozygosity occurs in the entire long arm or short arm. The heterozygosity distribution of each SNP in the East Asian population refers to the gnomAD_exome_EAS database. The selection criteria for SNPs are: the probability of the SNP being heterozygous is close to 50% in the population to avoid the situation that all SNPs are homozygous; the distance between SNPs in the genome is greater than 300kb to minimize the linkage bet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com