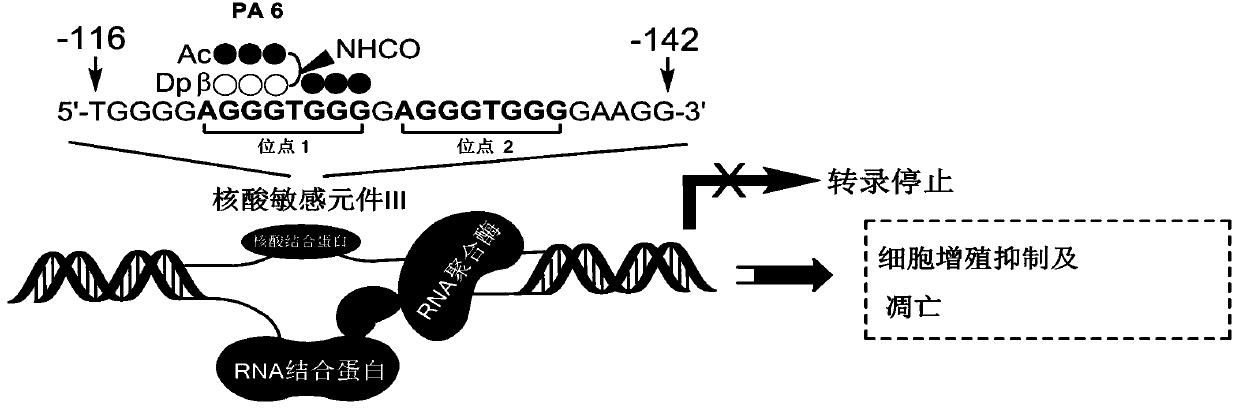
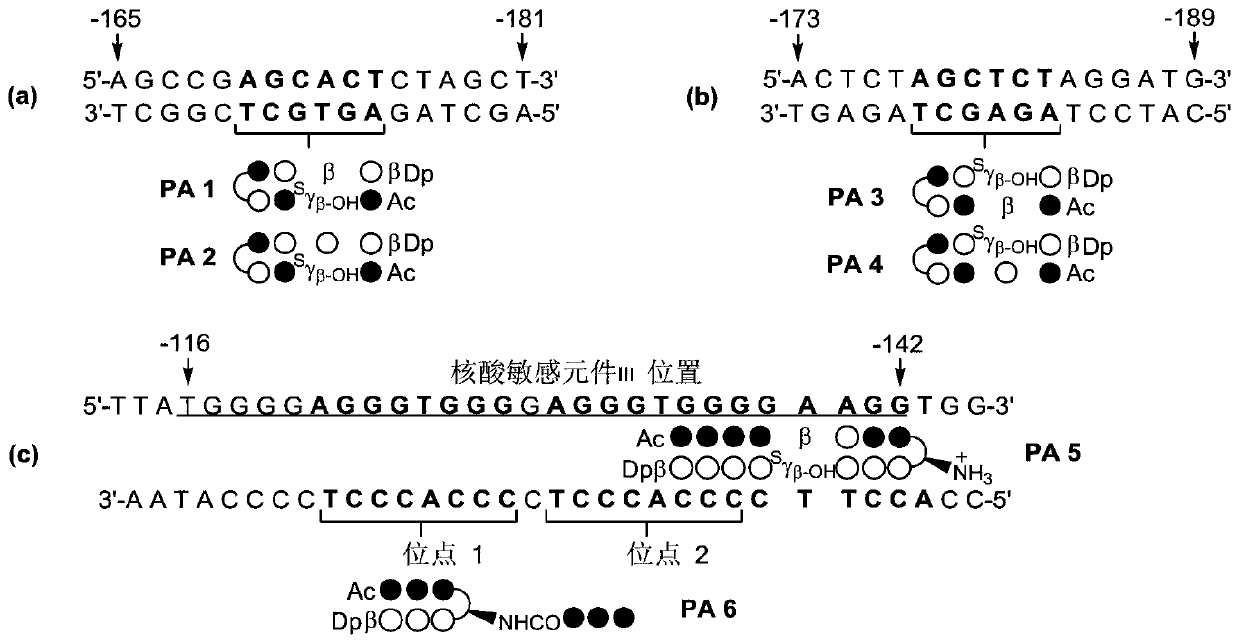
c-Myc gene expression inhibitor and application thereof
A gene expression, c-myc technology, applied in medical preparations containing active ingredients, organic active ingredients, drug combinations, etc. Toxicity, inducing tumor cell apoptosis, and inhibiting growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
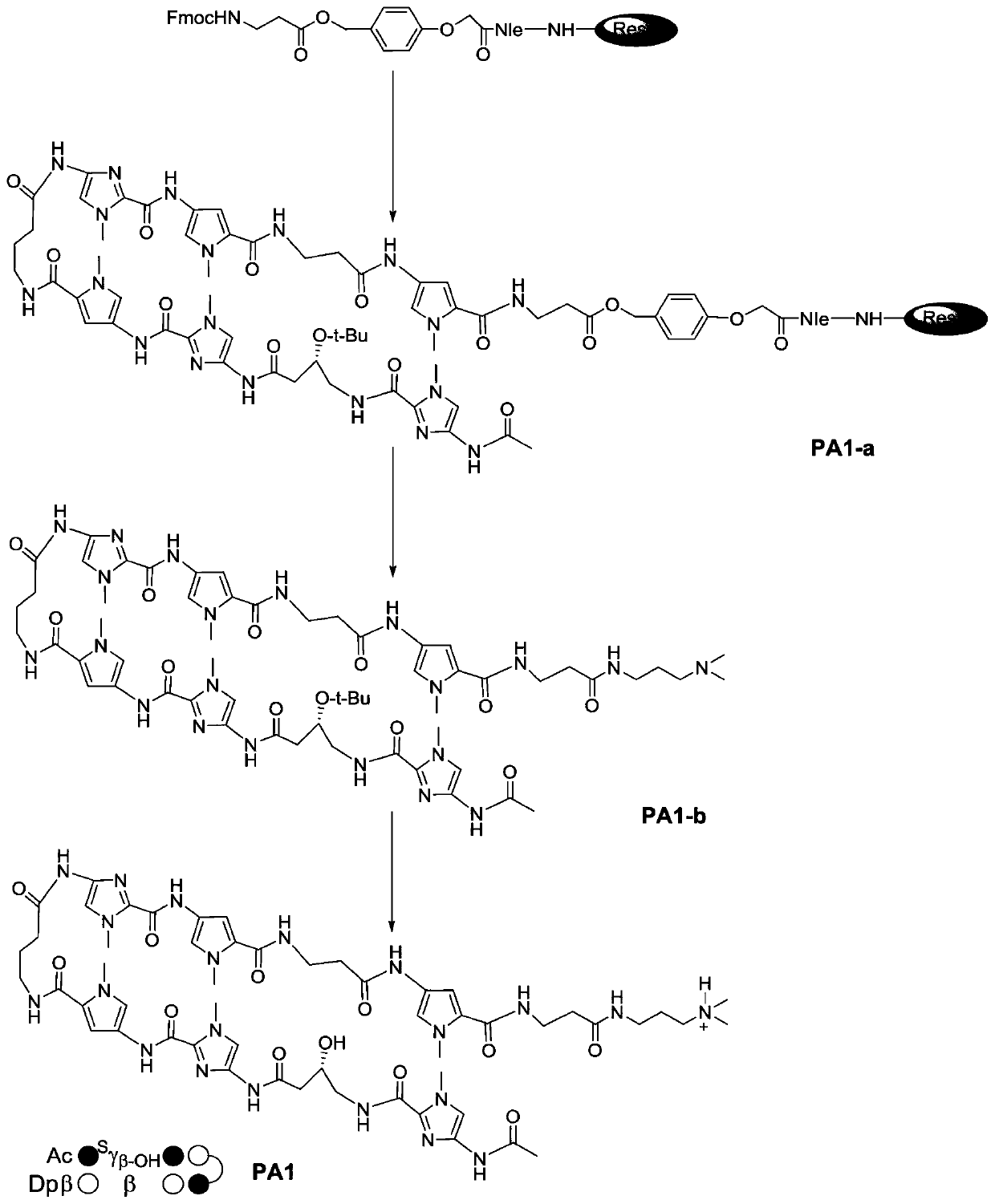
[0052] Embodiment 1: the preparation of compound PA1
[0053] The main synthesis steps: activate the resin, remove the protective group on the resin, activate the carboxyl group, couple, cap, cut the resin and other parts.
[0054] Activation of the resin: fully swell the resin with DMF.
[0055] Deprotection: Deprotect the Fmoc protecting group from the Fmoc-protected resin by piperidine / DMF solution to release the free amino group.
[0056] Activation of the carboxyl group: The carboxyl group of the amino acid involved in the next coupling reaction is activated by the activation reagent HBTU to form an activated ester, thereby efficiently coupling with the amino group.
[0057] Coupling: Activated amino acids react with free amino groups on the resin to form peptide bonds.
[0058] Capping: There are some incompletely reacted amino structures in the reaction, which must be capped to avoid side reactions in the next coupling, so as to ensure the purity of the synthesized ta...
Embodiment 2
[0092] Embodiment 2: the preparation of compound PA2
[0093] The synthesis steps are similar to the synthesis of PA1( Figure 3A ). Proceed as follows:
[0094] 1) Activation: same as the first step 1) activation of PA 1 preparation. weigh Figure 3B Fmoc protection β-alanine-Clear resin (SPS-1, 1.00g, 0.4mmol, Peptides International) as shown in formula (12) joins in the solid-phase reaction tube of solid-phase reaction device, logical nitrogen protection, again Add 5mL of DMF to fully bubble nitrogen for 30min to activate the resin;
[0095] 2) Deprotection: first prepare 3 mL of 20% (v / v) piperidine / DMF (both piperidine and DMF are treated anhydrous solvents), and add it to the solid-phase reaction tube of step 1) under nitrogen protection, Fully bubbling reaction for 15 minutes, remove the amino protecting group on β-alanine, remove the solvent in the reaction tube, rinse twice with 3mL anhydrous dichloromethane, rinse once with 2mL anhydrous DMF, each time Drain th...
Embodiment 3
[0119] Embodiment 3: the preparation of compound PA3
[0120] The synthesis steps are similar to the synthesis of PA1( Figure 3A ). Proceed as follows:
[0121] 1) Activation: same as the first step 1) activation of PA 1 preparation. weigh Figure 3B Fmoc protection β-alanine-Clear resin (SPS-1, 1.00g, 0.4mmol, Peptides International) as shown in formula (12) joins in the solid-phase reaction tube of solid-phase reaction device, logical nitrogen protection, again Add 5mL of DMF to fully bubble nitrogen for 30min to activate the resin;
[0122] 2) Deprotection: first prepare 3 mL of 20% (v / v) piperidine / DMF (both piperidine and DMF are treated anhydrous solvents), and add it to the solid-phase reaction tube of step 1) under nitrogen protection, Fully bubbling reaction for 15 minutes, remove the amino protecting group on β-alanine, remove the solvent in the reaction tube, rinse twice with 3mL anhydrous dichloromethane, rinse once with 2mL anhydrous DMF, each time Drain th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com