Scn9a antisense oligonucleotides
A peptide nucleic acid and nucleobase technology, applied in the field of peptide nucleic acid derivatives, can solve the problems of unreported alternative splicing and achieve high binding affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
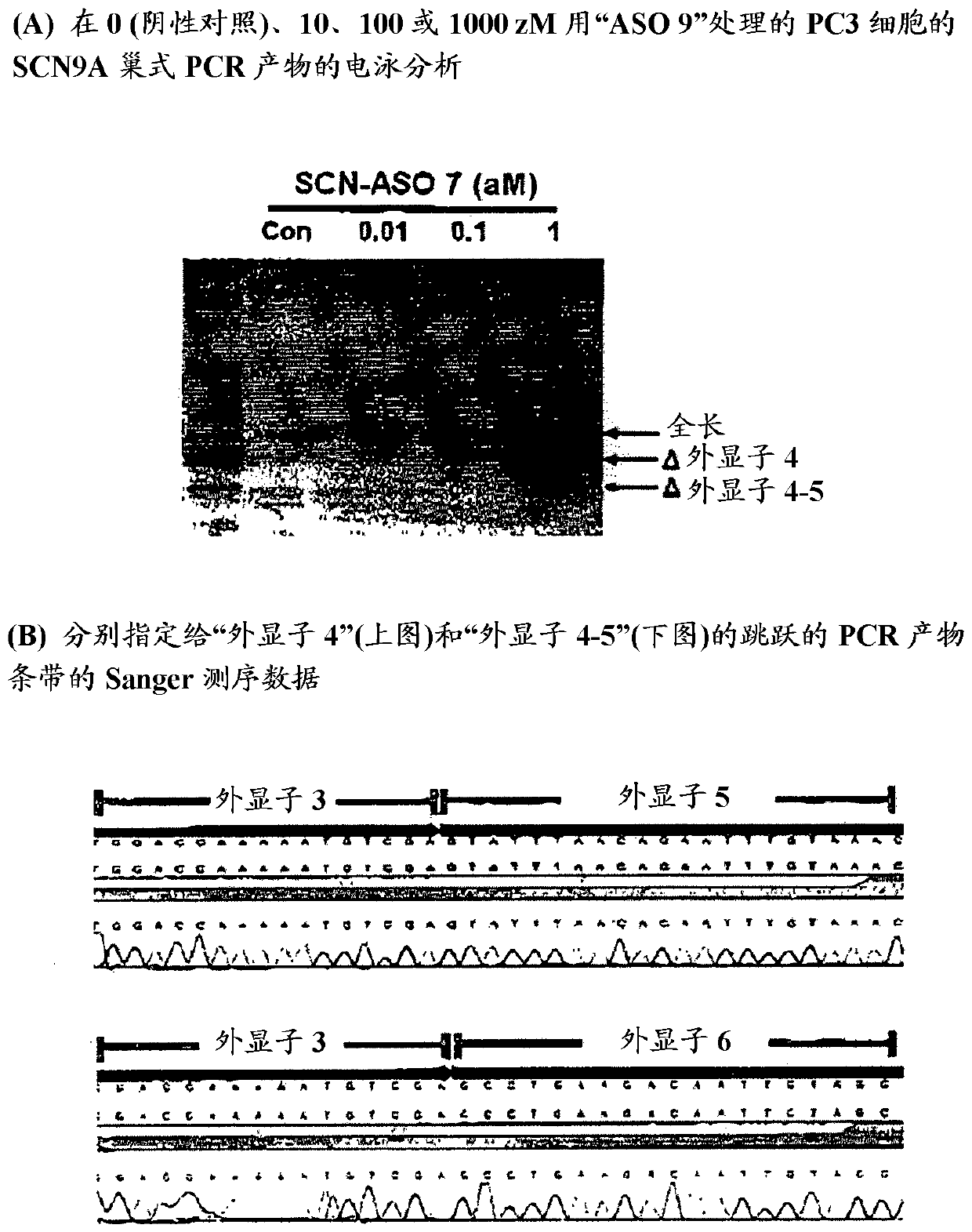
[0285] Example 1. Exon skipping induced by "ASO 9" in PC3 cells
[0286] 'ASO9' and [(5'→3')CGUCA spanning the junction of 'exon 4' and 'intron 4' in the human SCN9A pre-mRNA UUGUUUUUGC ┃ guaagu The 16-mer pre-mRNA sequences marked "bold" and "underlined" in the 30-mer sequence of acuuucagc] are complementary bound. "ASO 9" has a 10-mer overlap with "Exon 4" and a 6-mer overlap with "Intron 4". Thus, "ASO 9" satisfies the complementarity overlap criterion for compounds of formula I of the present invention.
[0287] Given the known high expression of human SCN9A mRNA in PC3 cells [ Br. J. Pharmacol vol 156, 420-431 (2009)], the ability of "ASO 9" to induce "exon 4" skipping of human SCN9A pre-mRNA in PC3 cells was assessed by SCN9A nested RT-PCR as described below.
[0288] [Cell culture and ASO treatment] Make PC3 cells (Cat. No. CRL-1435, ATCC) in 5 mL of Ham's F-12K medium supplemented with 10% FBS, 1% streptomycin / penicillin, 1% L-glutamine amide and 1% sodium pyru...
Embodiment 2
[0293] Example 2. qPCR of SCN9A mRNA in PC3 cells treated with "ASO 9"
[0294] The ability of "ASO 9" to induce changes in the expression level of human SCN9A mRNA in PC3 cells was assessed by qPCR against an exon-specific primer set covering "Exons 4-6" as follows.
[0295] [Cell culture and ASO treatment] PC3 cells grown in a 60 mm dish containing 5 mL of F-12K medium were incubated with "ASO 9" at 0 (negative control), 10, 100 or 1000 zM for 24 hours. (2 Petri dishes for each ASO concentration).
[0296] [RNA Extraction] Using "MiniBEST Universal RNA Extraction Kit" (Cat. No. 9767, Takara), total RNA was extracted according to the manufacturer's instructions.
[0297] [Synthesis of cDNA by one-step RT-PCR] Using Super Script ® One-Step RT-PCR Kit and Platinum ® Taq polymerase (Catalog No. 10928-042, Invitrogen) and a set of exon-specific primers [exon 2_forward: (5'→3') CTTTCTCCTTTCAGTCCTCT; and exon 9_reverse: ( 5'→3')TTGCCTGGTTCTGTTCTT], according to the following cy...
Embodiment 3
[0300] Example 3. Inhibition of sodium current in PC3 cells treated with "ASO 9"
[0301] Cellular sodium currents are usually measured by patch clamp. As sodium ions enter cells, intracellular sodium ion levels increase. Intracellular sodium levels can be detected using sodium ion-sensitive dyes. "CoroNa Green" is a dye with a crown ether type sodium ion chelating agent. "CoroNa Green" emits green fluorescence when it chelates sodium ions. "CoroNa Green" has been used to indirectly measure intracellular sodium levels. Sodium levels measured by "CoroNa Green" were found to correlate well with sodium ion currents measured by sodium ion patch clamp[ Proc. Natl. Acad. Sci. USA vol 106(38), 16145-16150 (2009)].
[0302] PC3 cells are known to express abundant human SCN9A mRNA and sodium currents, despite co-expressing other SCN isoforms[ Br. J. Pharmacol . vol 156, 420-431 (2009)]. Therefore, if Na v The sodium ion current of 1.7 sodium channel subtypes accounts for a si...
PUM
| Property | Measurement | Unit |
|---|---|---|
| fluorescence | aaaaa | aaaaa |
| fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com