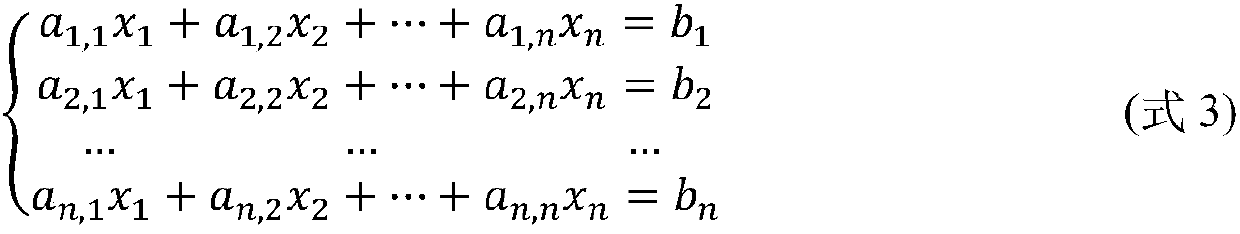Colony parallel computation accelerating method for calculating protein structure based on protein folding
A protein folding and parallel computing technology, which is applied in the field of high-performance computing, can solve the problems of large-scale stand-alone capacity, low degree of parallelization, and unsatisfactory acceleration effects, etc., to improve the calculation solution speed and calculation accuracy, improve the calculation speed, Effect of reducing communication overhead
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0047] Below in conjunction with accompanying drawing, further describe the present invention through embodiment, but do not limit the scope of the present invention in any way.
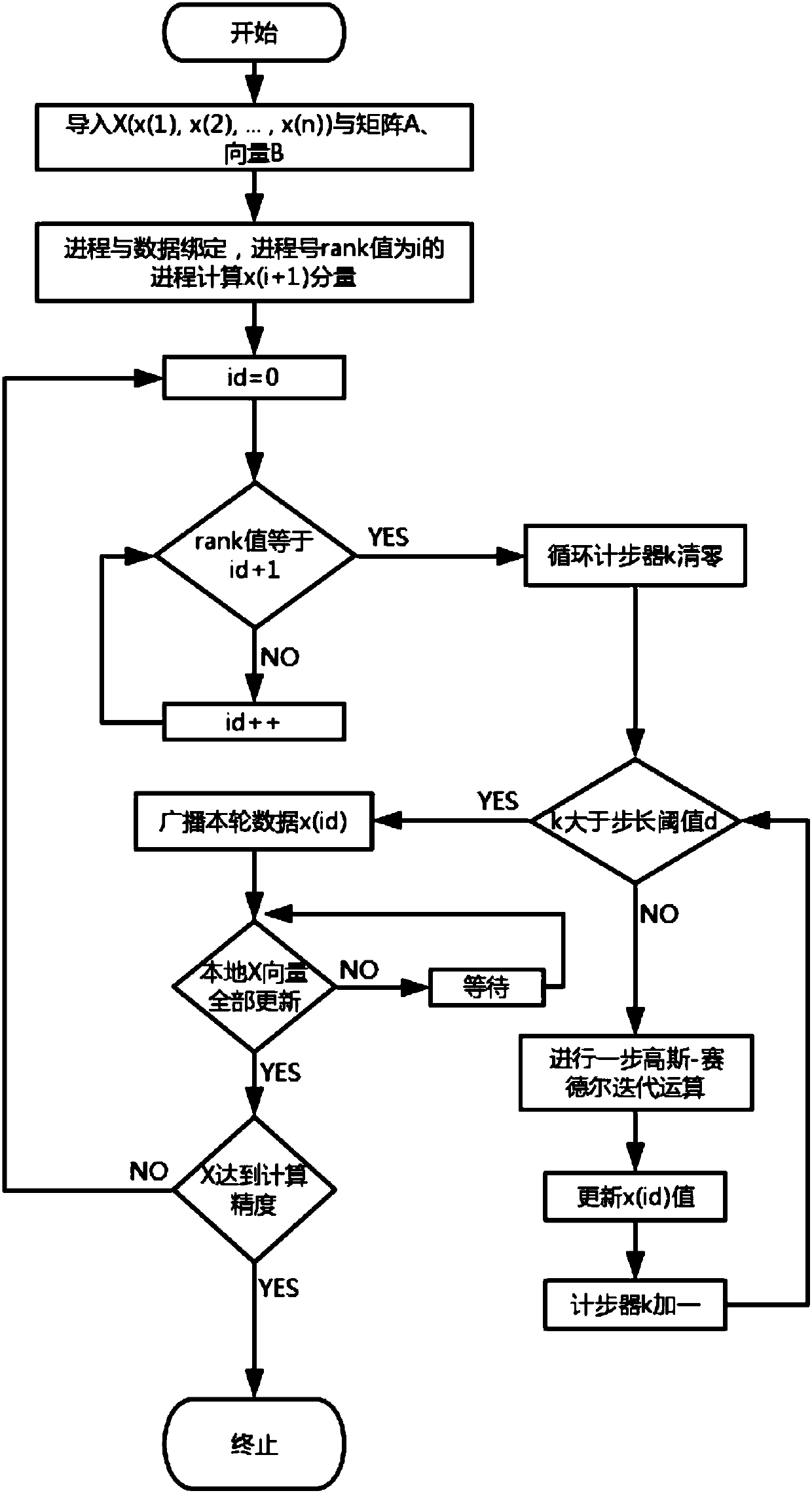
[0048] The present invention provides a parallel acceleration scheme for the protein folding problem on a cluster. After the method of the present invention constructs the energy-constrained linear equations of protein folding, the linear equations are divided into reasonable blocks, and the calculation is parallelized and accelerated on the cluster, and the cluster parallelization of the linear equations provided by the present invention The acceleration method improves the calculation speed of the iterative solution of linear equations, reduces the communication overhead in the calculation process, and saves a lot of computing resources, thereby improving the calculation and solution speed and calculation accuracy of protein structure calculation. figure 1 It is a flow chart of the DSP computing m...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap