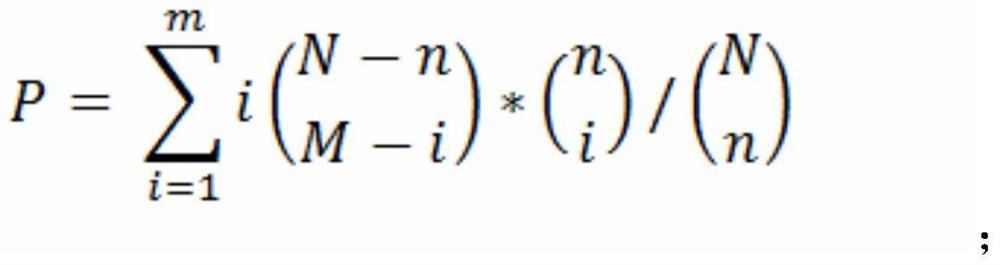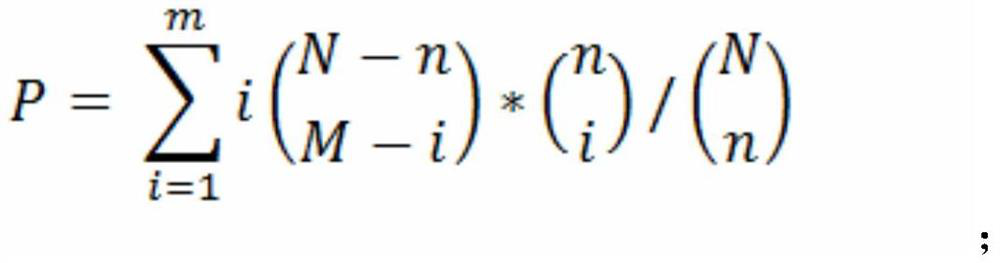A method for joint analysis of multi-omics data
A technology of omics data and joint analysis, applied in the field of bioinformatics, can solve problems such as high method limitations, poor versatility, and difficulty in meeting scientific research needs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1, joint analysis of rat mRNA and small RNA data
[0057] 1. Experimental samples
[0058] Normal tissue samples from rats at 6 different developmental time stages.
[0059] 2. Data collection
[0060] Using conventional RNA-Seq and small RNA standard information analysis, the input data of mRNA and small RNA were obtained from normal tissue samples of 6 different developmental time stages of rats.
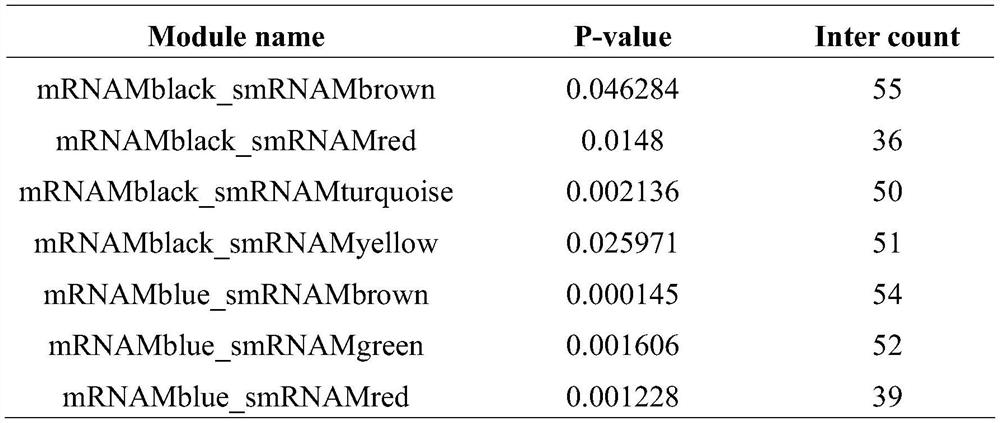
[0061] Finally, the mRNA data includes the expression levels of 25,289 genes, and the small RNA data includes the expression levels of 631 miRNAs. The targeting relationship between miRNA and mRNA is based on the prediction results of TargetScan and miRanda target gene prediction software, including 76069 pairs of targeting relationships.
[0062] 3. Data preprocessing
[0063] Delete data with a missing rate higher than the set threshold (the default threshold is 0.2, that is, the missing rate is 20%) (delete the corresponding rows).
[0064] 4. Co-expressi...
Embodiment 2
[0097] Example 2, joint analysis of apple methylation, mRNA and small RNA data
[0098] 1. Experimental samples
[0099] Samples of 4 different treatments of apples.
[0100] 2. Data collection
[0101] The input data of mRNA, methylation and small RNA were obtained from 4 different apple samples by routine RNA-Seq, genome-wide Bisulfite methylation and Small RNA standard information analysis.
[0102] Finally, the data of mRNA include the expression of 31,964 genes, the data of small RNA are the expression of 167 miRNAs, and the methylation is the result of the methylation rate of 34,889 methylated regions. The targeting relationship between miRNA and mRNA is based on the prediction results of psRobot and targetfinder target gene prediction software, including 9033 pairs of targeting relationships.
[0103] 3. Data preprocessing
[0104] Delete data with a missing rate higher than the set threshold (the default threshold is 0.2, that is, the missing rate is 20%) (delete t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com