Kit and system for prognosis prediction of metastatic colorectal cancer
A colorectal cancer and prediction system technology, applied in the field of biomedicine, can solve problems such as the limitations of molecular heterogeneity in metastatic colorectal cancer, and achieve good prediction results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Construction of a prognosis prediction model for metastatic colorectal cancer
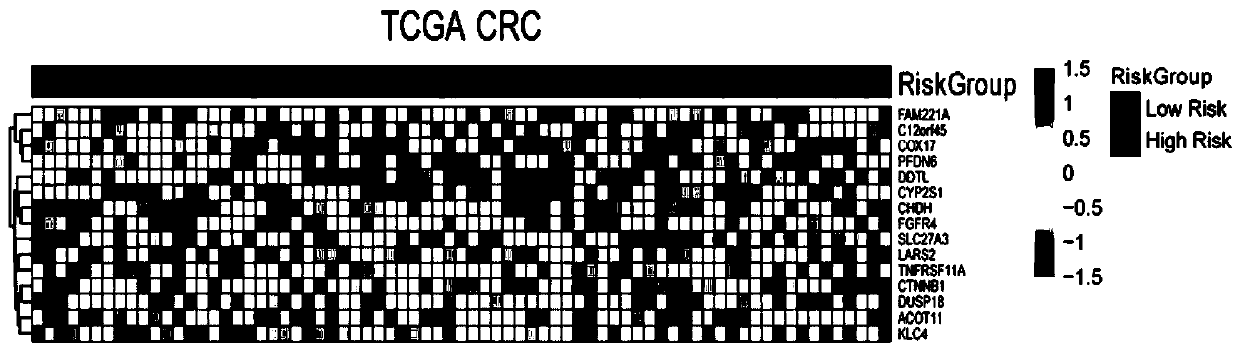
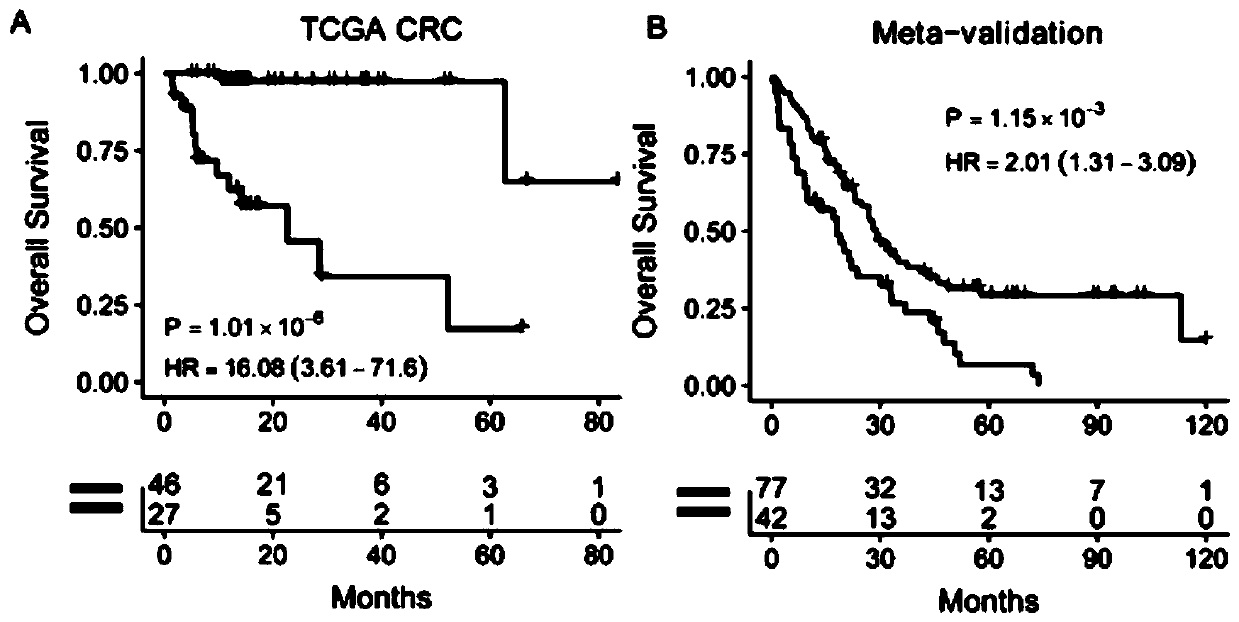
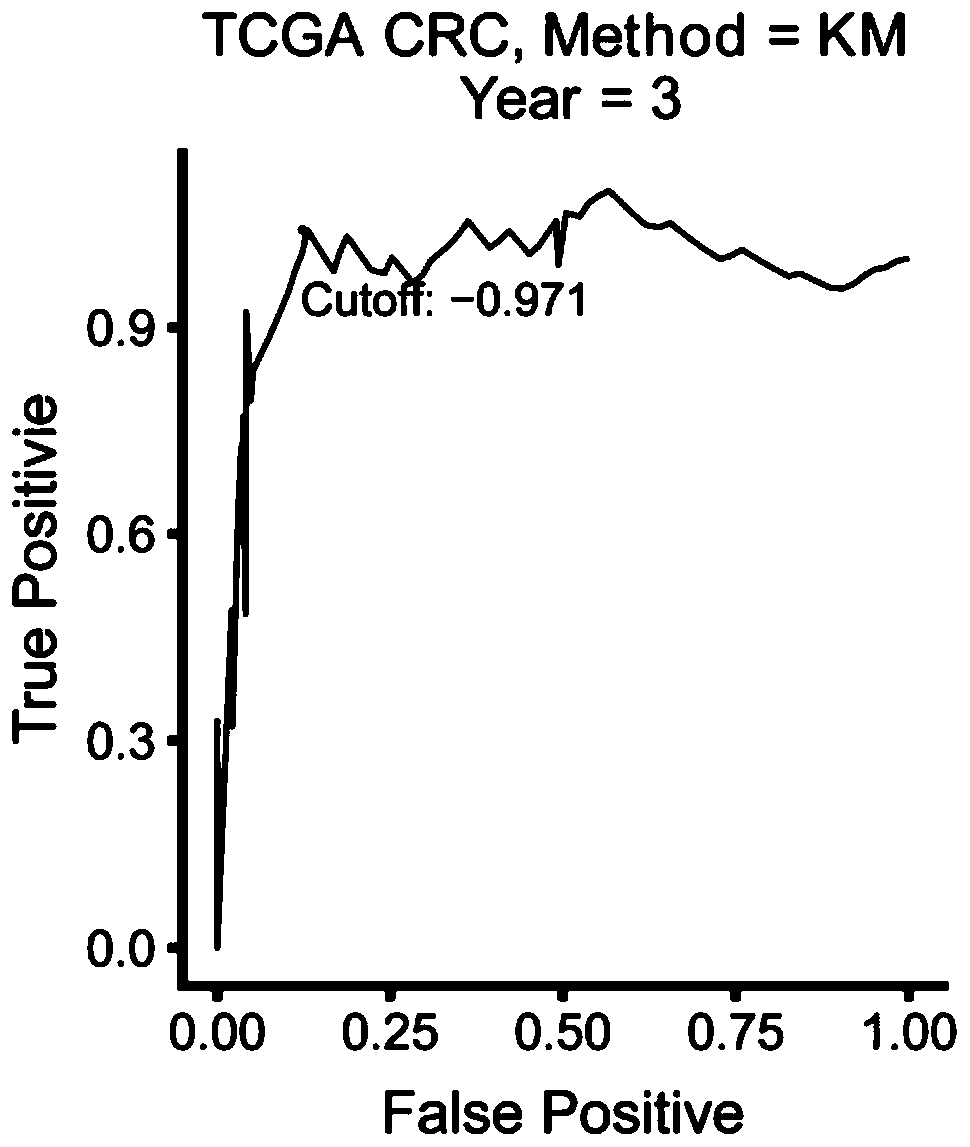
[0045] (1) Gene expression profiles of frozen colorectal cancer tumor tissue samples from 4 public cohorts were analyzed, including datasets from TCGA CRC and 3 microarray datasets (GSE39582, GSE39084, and GSE17536) obtained from Gene Expression Omnibus . In this example, 192 metastatic colorectal cancer patients with valid survival information were included, and 18113 genes were analyzed. First, 4172 genes were obtained by filtering the median absolute deviation, that is, Median Absolute Deviation greater than 0.5. Further sampling and analysis were carried out by using sampling technology, and 197 prognostic genes were obtained. Finally, a prognostic gene signature (PGS) consisting of 15 genes associated with the prognosis of metastatic colorectal cancer was constructed using the LASSO Cox model. The 15 genes associated with the prognosis of metastatic colorectal cancer are: ACOT11, C12...
Embodiment 2
[0051] Prediction of Prognosis in Metastatic Colorectal Cancer
[0052] (1) qPCR reaction detection
[0053] Detect the expression levels of the following target genes in patients with metastatic colorectal cancer to be predicted, that is, the Ct value of mRNA: ACOT11, C12orf45, CHDH, COX17, CTNNB1, CYP2S1, DDTL, DUSP18, FAM221A, FGFR4, KLC4, LARS2, PFDN6, SLC27A3 and TNFRSF11A.
[0054] Use qPCR technology to detect the expression level of the target gene:
[0055] Use the primer pairs of the genes in the following table 1 to detect the expression level of the corresponding genes
[0056] Table 1 Primer sequences for detection of genes related to prognosis of metastatic colorectal cancer
[0057]
[0058]
[0059] a) qPCR reaction system:
[0060] Prepare the mixture of template RNA and primers as follows:
[0061]
[0062] *The reaction system can be scaled up
[0063] *Oligo(dT)15Primer and Random Primer are used at the same time, which can effectively improv...
Embodiment 3
[0080] This embodiment provides a metastatic colorectal cancer prognosis prediction device, the prediction device includes: a gene expression level information acquisition module, a risk value calculation module and a prediction module;
[0081] The gene expression level information acquisition module is used to obtain the expression level information of the target gene in patients with metastatic colorectal cancer.
[0082] Among them, the target genes include the following genes:
[0083] ACOT11, C12orf45, CHDH, COX17, CTNNB1, CYP2S1, DDTL, DUSP18, FAM221A, FGFR4, KLC4, LARS2, PFDN6, SLC27A3, and TNFRSF11A.
[0084] The risk value calculation module is used to substitute the expression level information of the target gene into the prediction model to calculate the risk value;
[0085] The calculation formula of the prediction model is as follows:
[0086] Risk value=-0.073×expACOT11+0.152×expC12orf45-0.179×expCHDH+0.518×expCOX17-0.054×expCTNNB1-0.065×expCYP2S1-0.001×expDDT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com