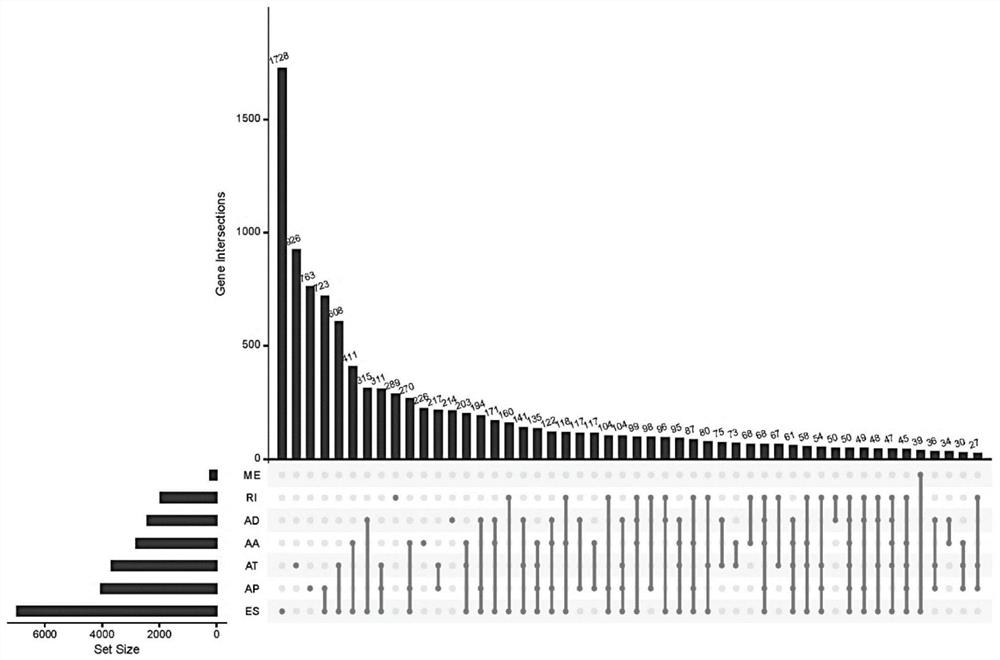
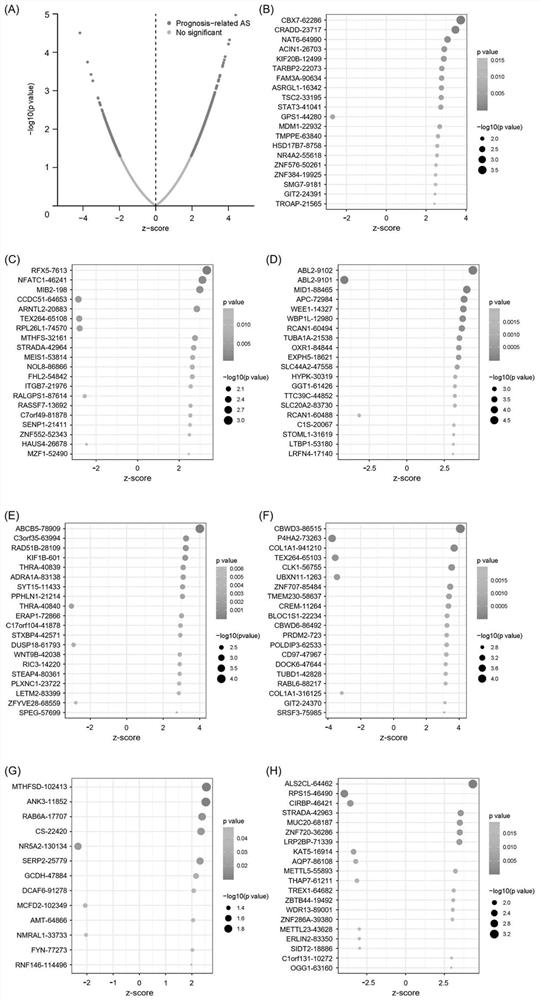
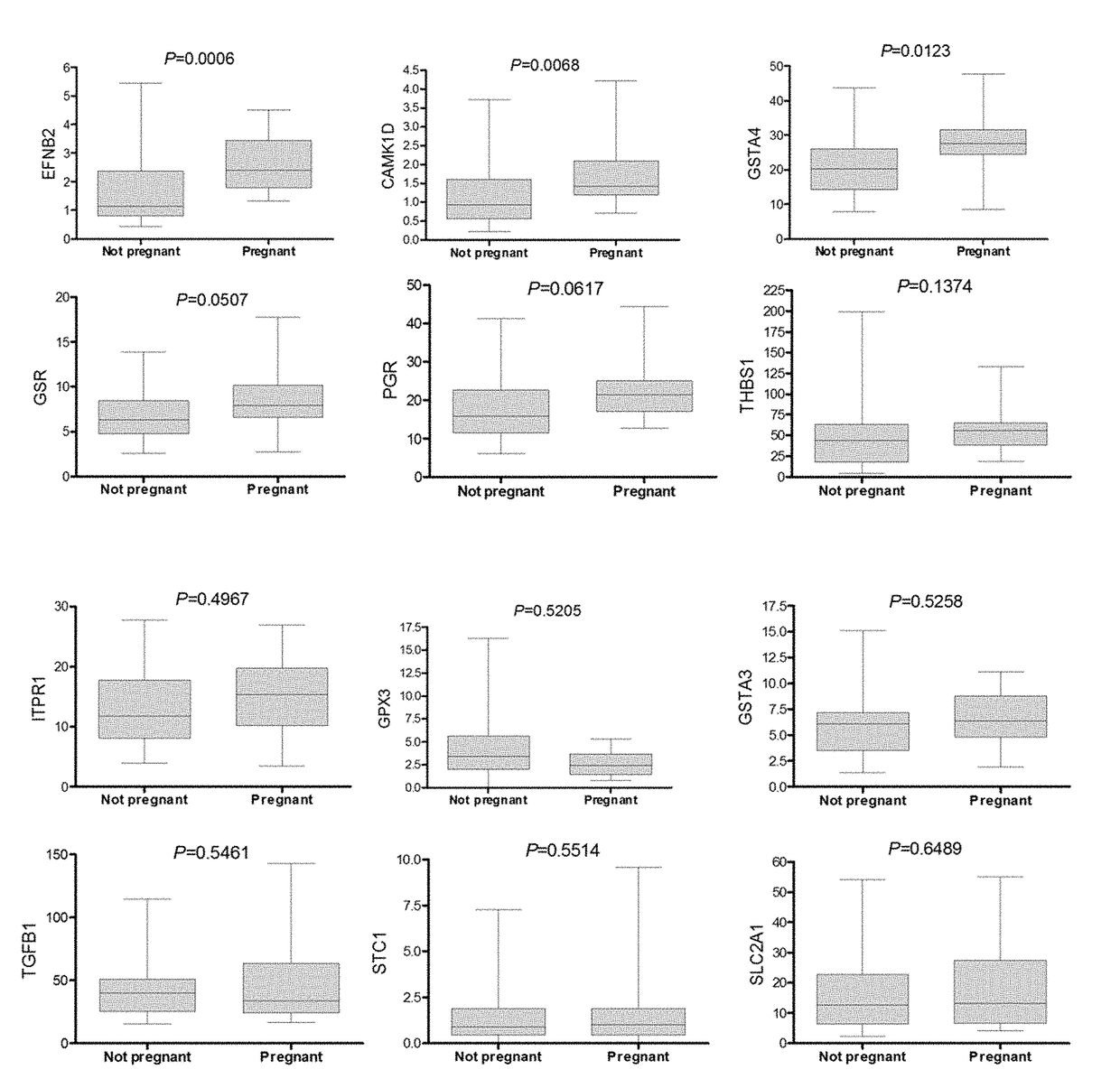
Patents
Literature
42 results about "Prognostic models" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Prognostic models are statistical tools that predict a clinical outcome based on at least 2 points of patient data. 2. Prognostic models are based on prognostic information that generally addresses the patient rather than the disease or treatment.
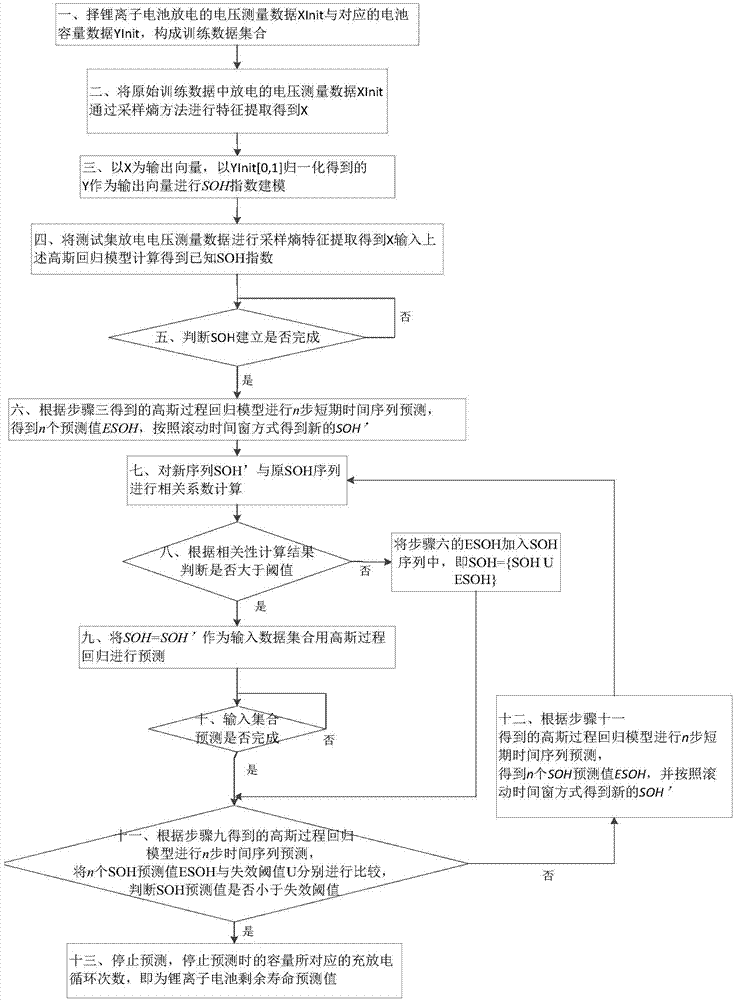
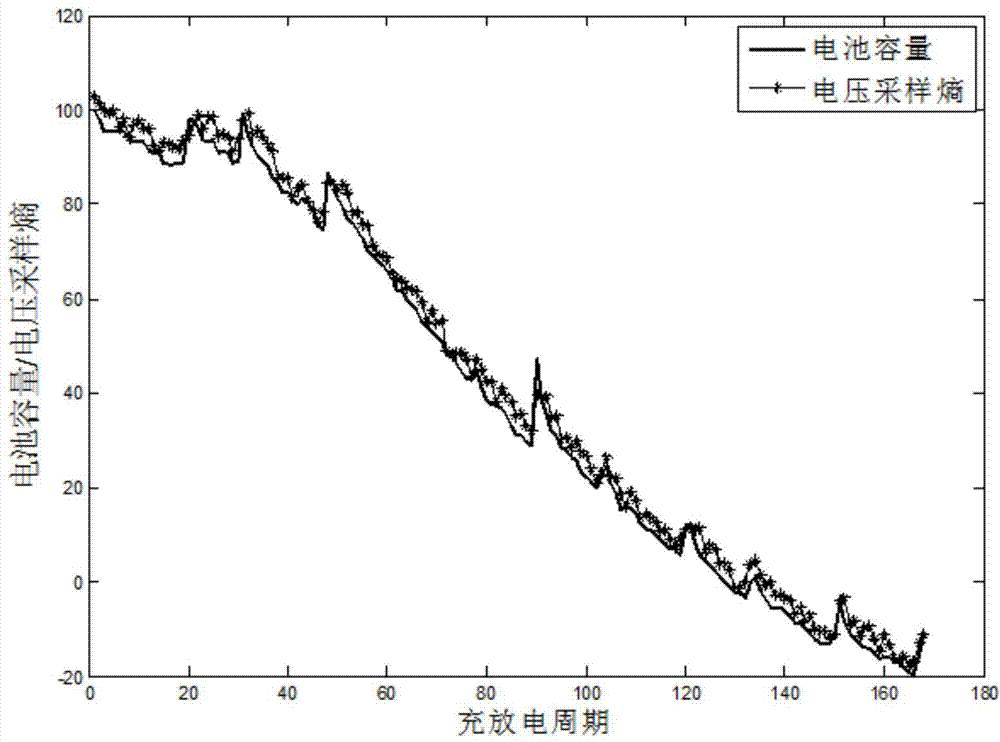
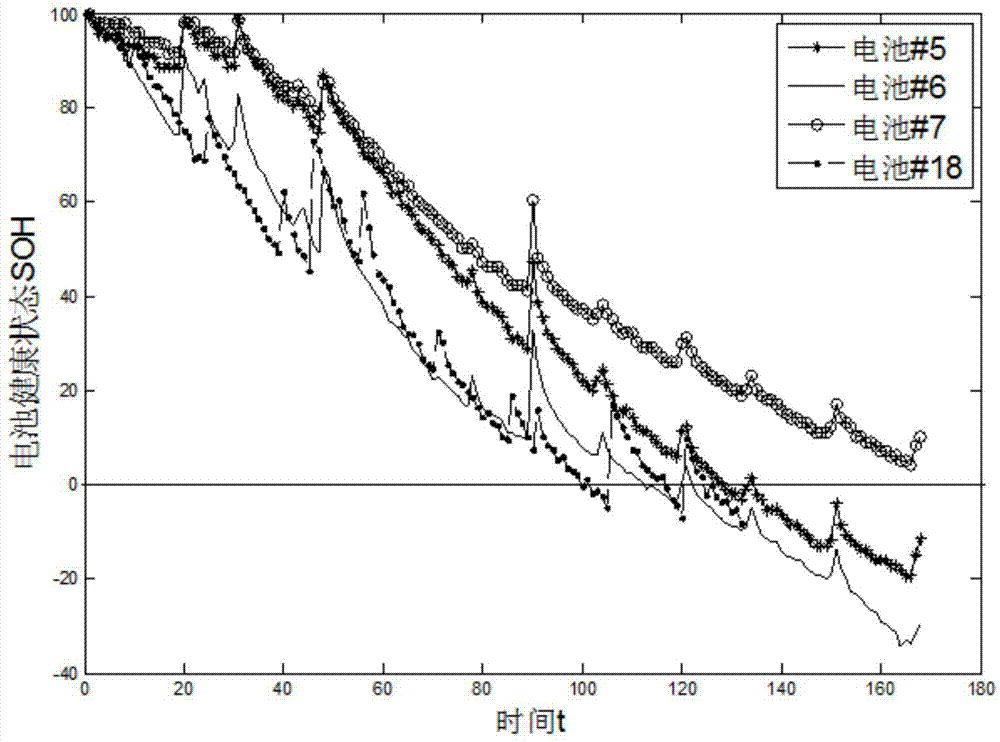
Method for remain useful life prognostic of lithium ion battery with model active updating strategy
InactiveCN103778280AEasy Adaptive AcquisitionFlexible inferenceElectrical testingSpecial data processing applicationsHealth indexEngineering
The invention relates to a method for remain useful life prognostic of a lithium ion battery with a model active updating strategy. According to a time series obtained through a voltage range of a discharge curve, conversion is conducted so that an equivalent discharge difference series obtained by discharge circulation at each time can be obtained, and therefore a health index time series of the ion battery is obtained; according to correspondence of a discharge voltage series and a time series, prognostic is conducted on the health index series to determine the remain useful life of the battery. Sampling entropy characteristic extraction and modeling are conducted on a charge voltage curve so that a relationship between a complete and accurate charge / discharge process and a battery performance index can be provided. On the basis of a performance index model, a short-term time series prognostic result is continuously updated to a known performance index data series and correlation analysis is conducted. According to the difference of the correlation degrees, retraining is conducted in the mode of training set expansion. The method is different from an existing iteration updating draining method, the prognostic model is updated dynamically, and therefore the prognostic precision is improved.
Owner:SHANGHAI JIAO TONG UNIV
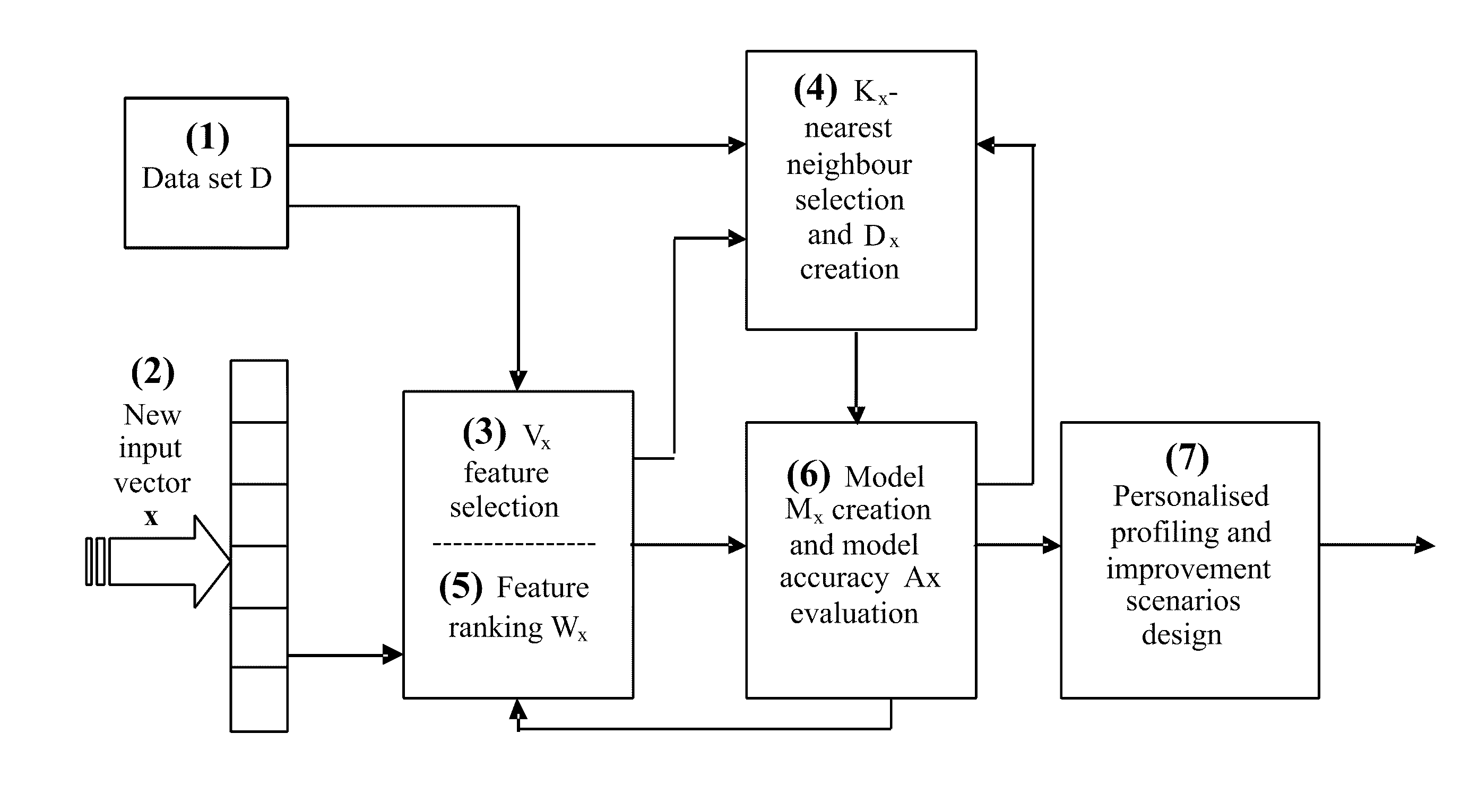
Data analysis and predictive systems and related methodologies
ActiveUS20110307228A1Maximum accuracyEasily realizedBiostatisticsForecastingData setVariable features
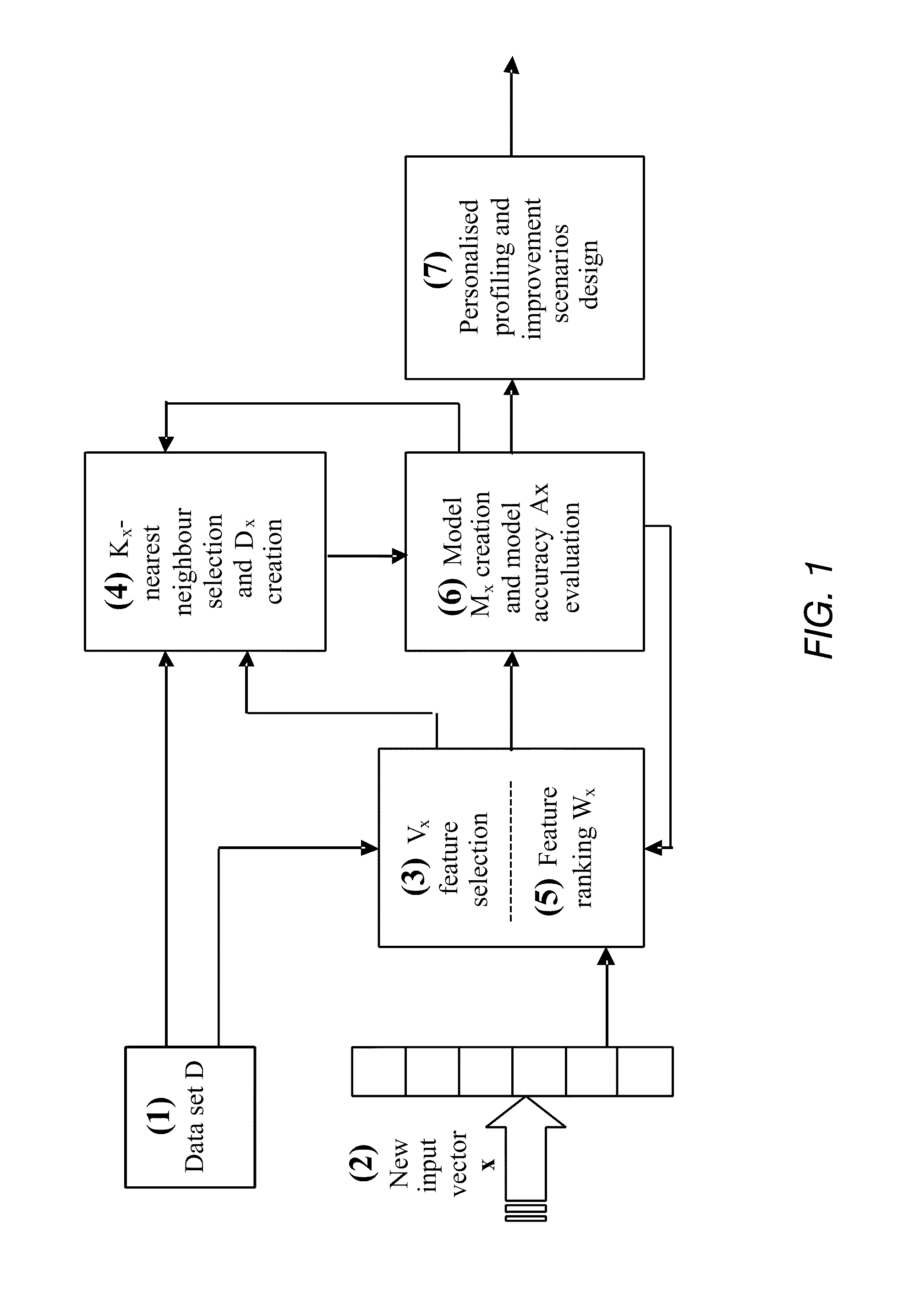
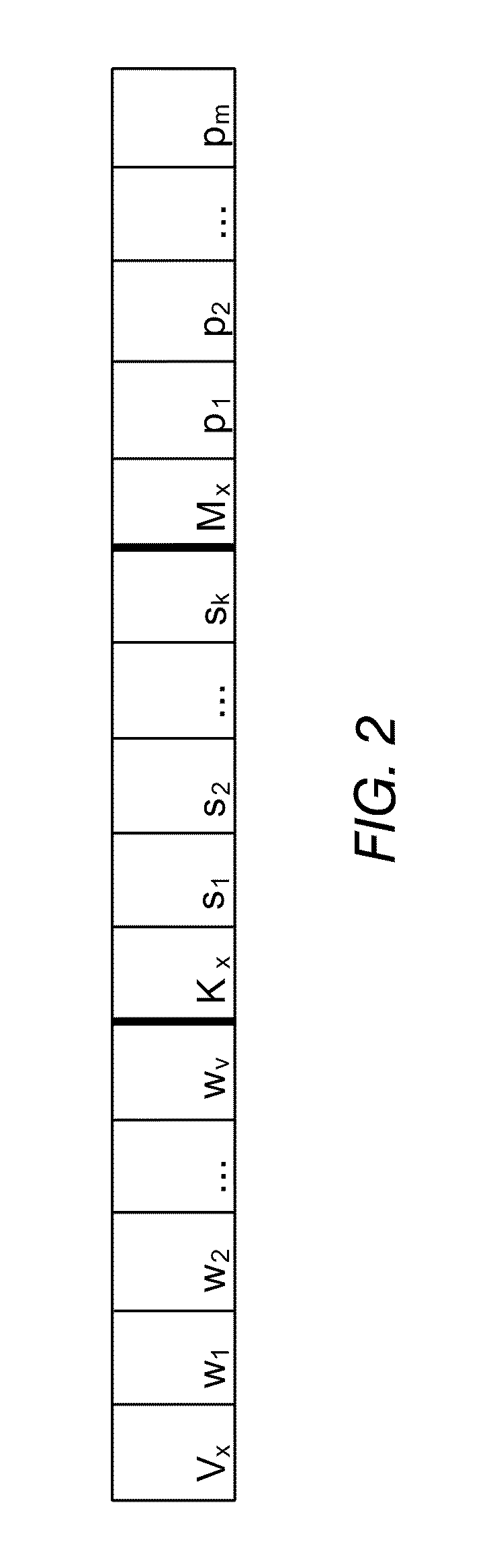
A method of optimising a model Mx suitable for use in data analysis and determining a prognostic outcome specific to a particular subject (input vector x), the subject comprising a number of variable features in relation to a scenario of interest for which there is a global dataset D of samples also having the same features relating to the scenario, and for which the outcome is known is disclosed. In one implementation, the method includes: (a) determining what number and a subset Vx of variable features will be used in assessing the outcome for the input vector x; (b) determining what number Kx of samples from within the global data set D will form a neighbourhood about x; (c) selecting suitable Kx samples from the global data set which have the variable features that most closely accord to the variable features of the particular subject x to form the neighbourhood Dx; (d) ranking the Vx variable features within the neighbourhood Dx in order of importance to the outcome of vector x and obtaining a weight vector Wx for all variable features Vx; (e) creating a prognostic model Mx, having a set of model parameters Px and the other parameters from (a)-(d); (f) testing the accuracy of the model Mx at e) for each sample from Dx; (g) storing both the accuracy from (f), and the model parameters developed in (a) to (e); (h) repeating (a) and / or (b) whilst applying an optimisation procedure to optimise Vx and / or Kx, to determine their optimal values, before repeating (c)-(h) until maximum accuracy at (f) is achieved.
Owner:KASABOV NIKOLA KIRILOV
Prognosis Modeling From One or More Sources of Information
ActiveUS20080033894A1Medical data miningDigital computer detailsPredictive modellingClinical psychology
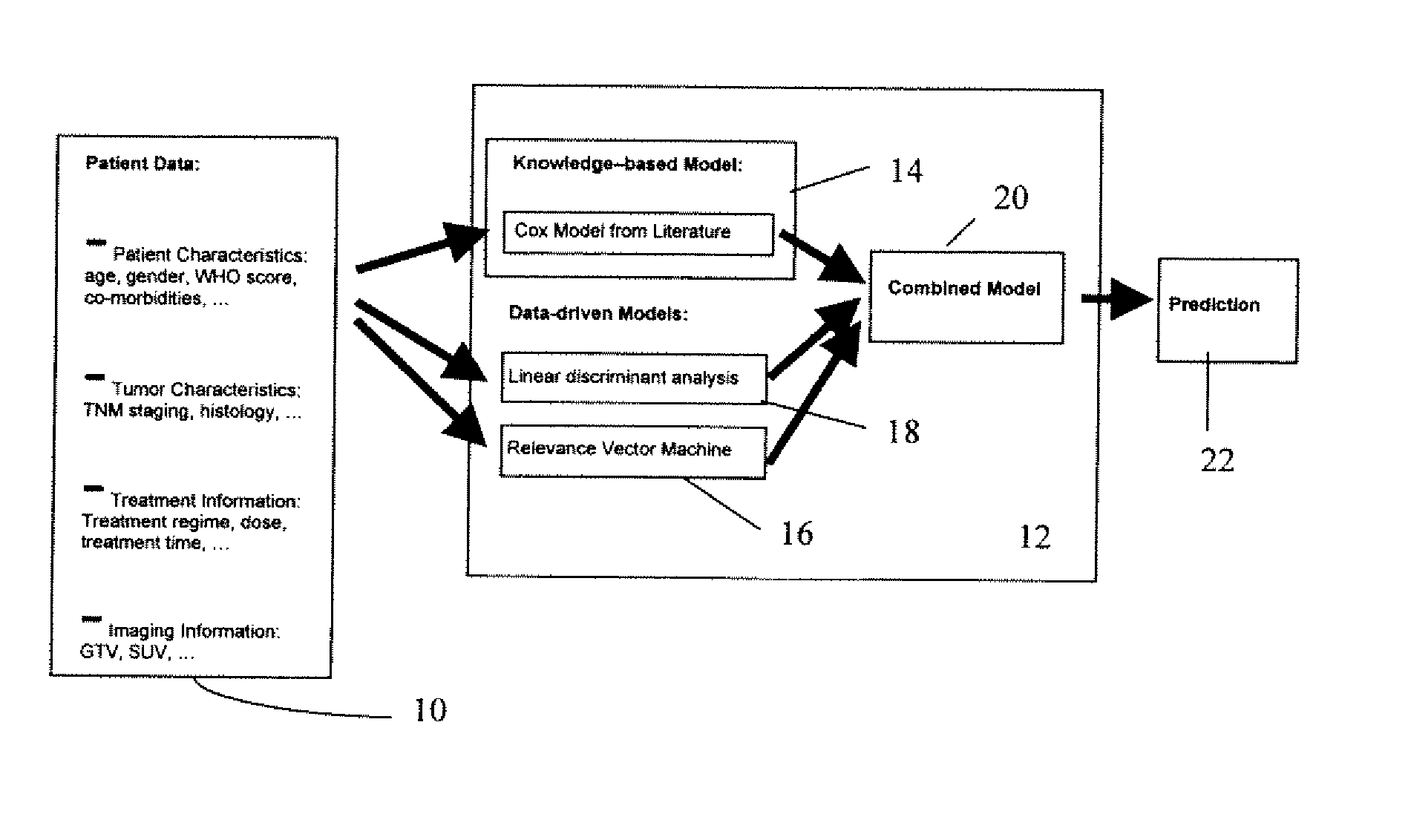
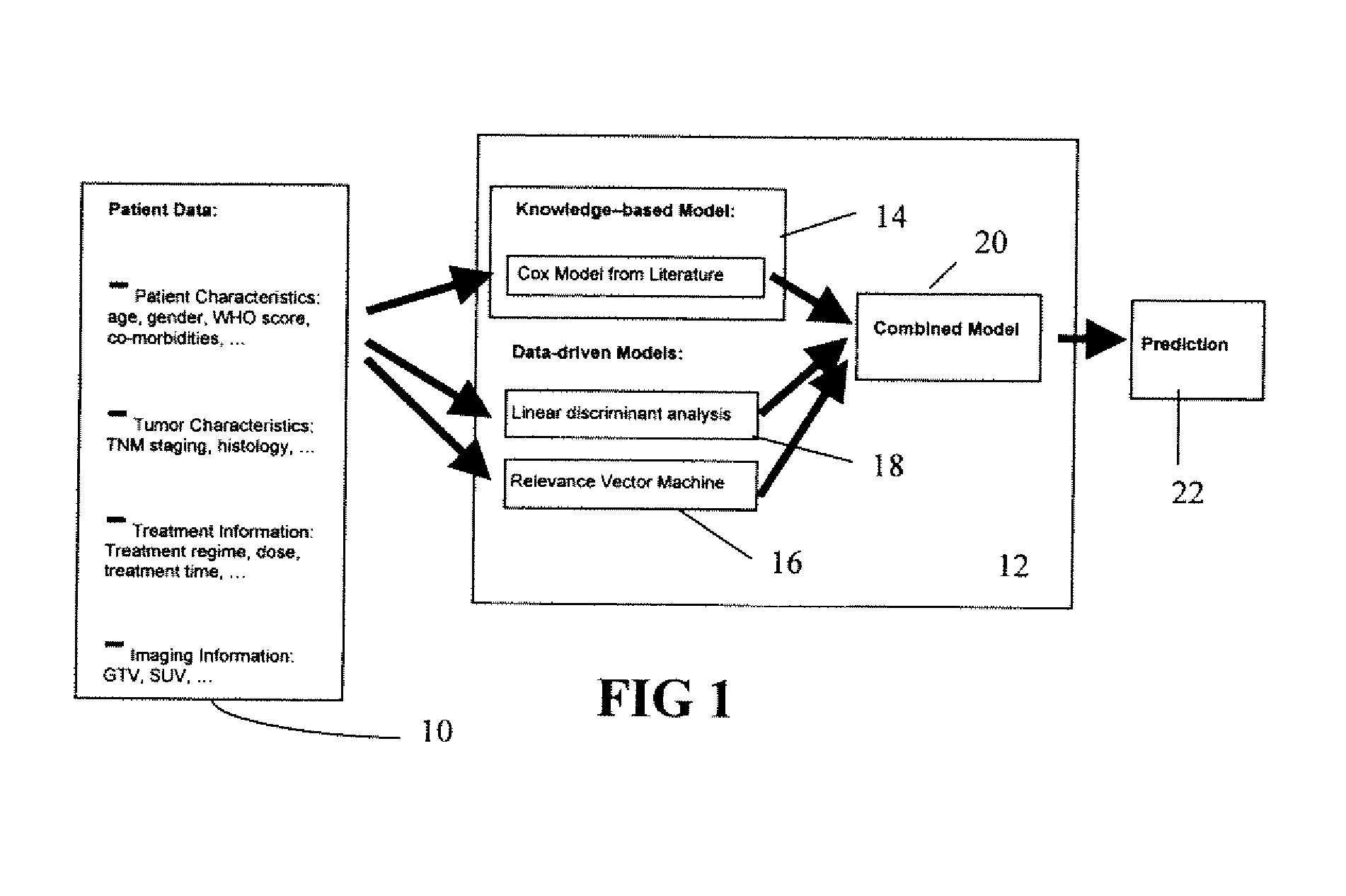
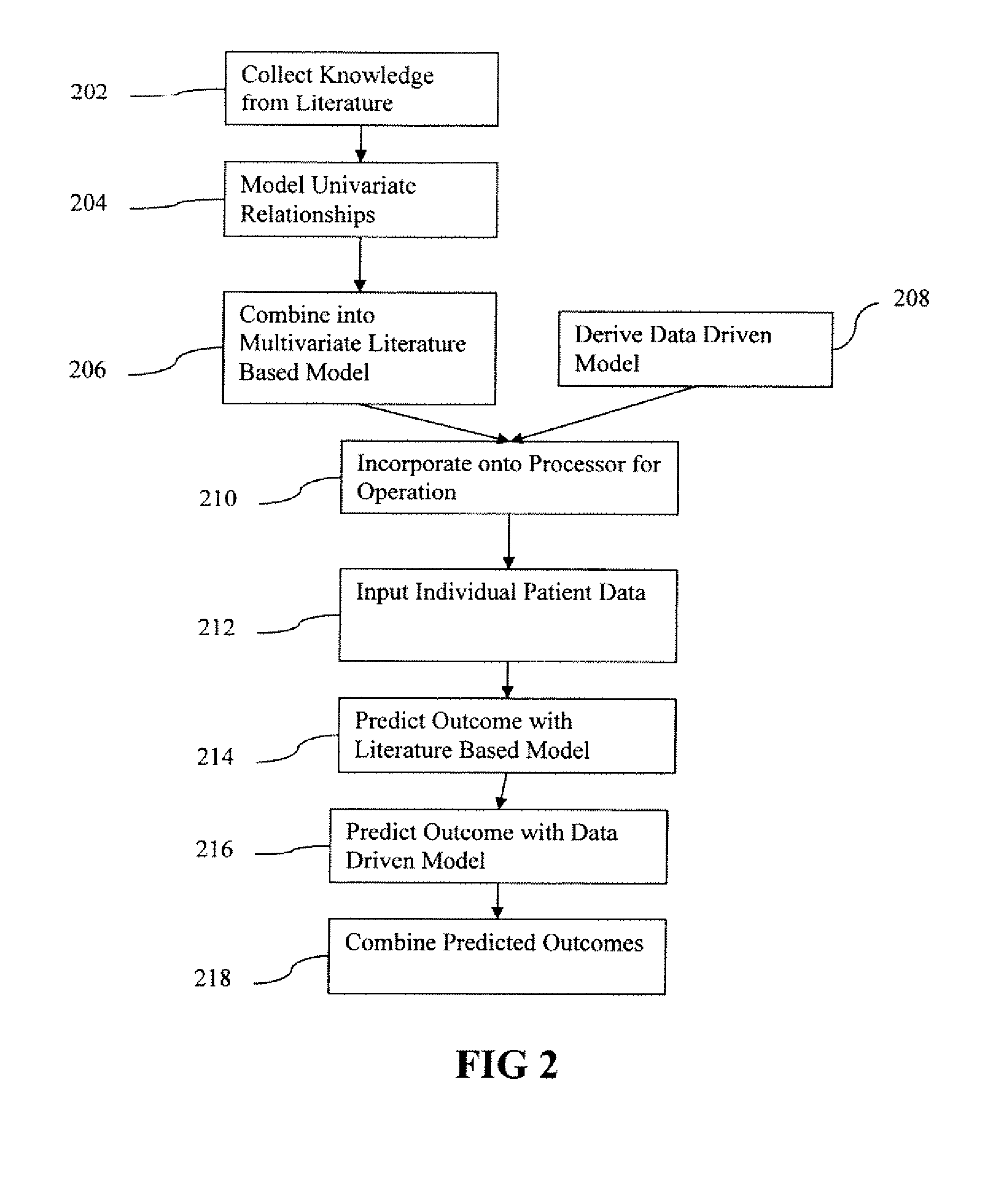
A predictor of medical treatment outcome is developed and applied. A prognosis model is developed from literature. The model is determined by reverse engineering the literature reported quantities. A relationship of a given variable to a treatment outcome is derived from the literature. A processor may then use individual patient values for one or more variables to predict outcome. The accuracy may be increased by including a data driven model in combination with the literature driven model.
Owner:CERNER INNOVATION
Diagnostic and Prognostic Tests
InactiveUS20090104617A1Simple powerEasy to adaptMicrobiological testing/measurementMaterial analysisTissue sampleNormal tissue
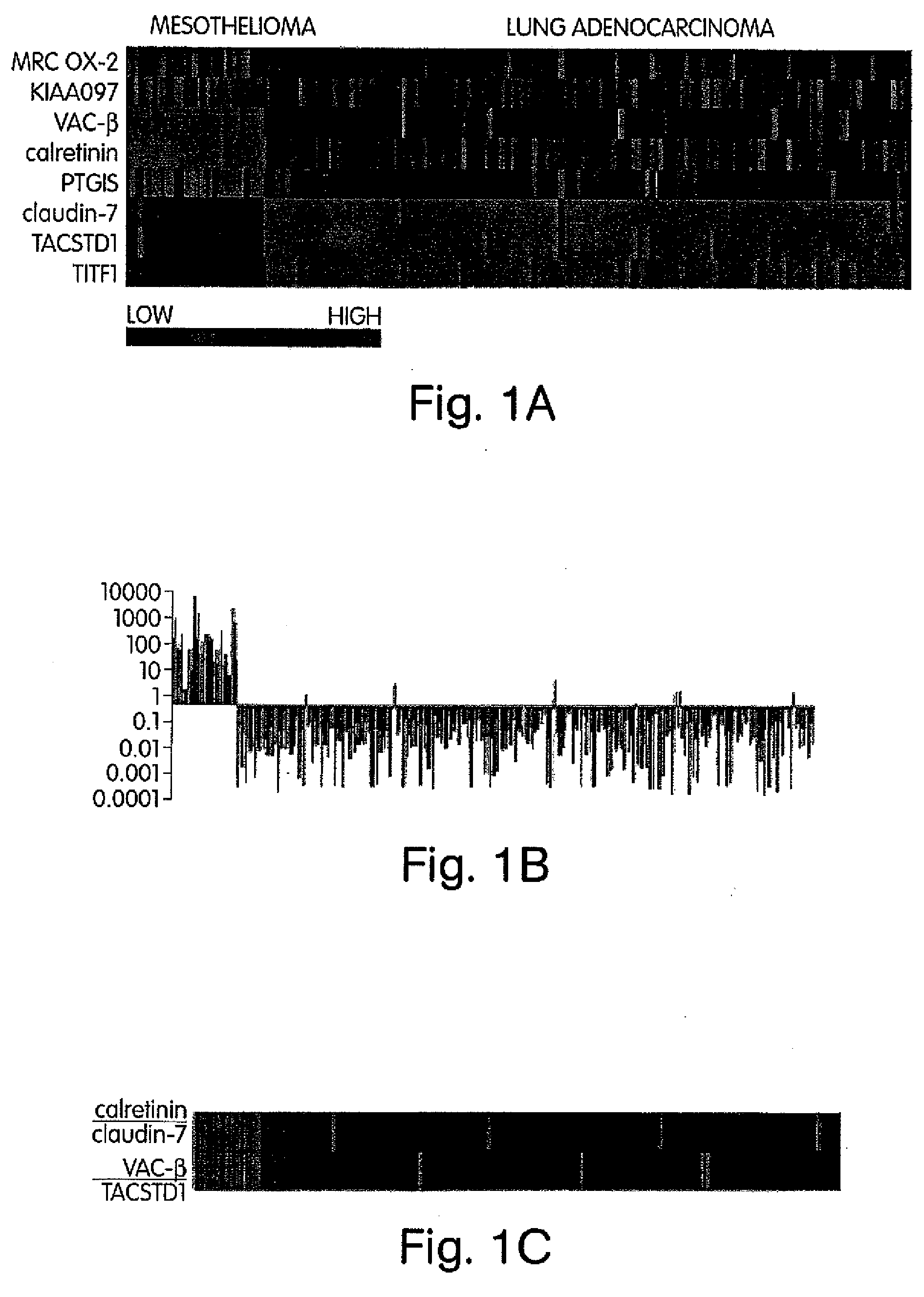
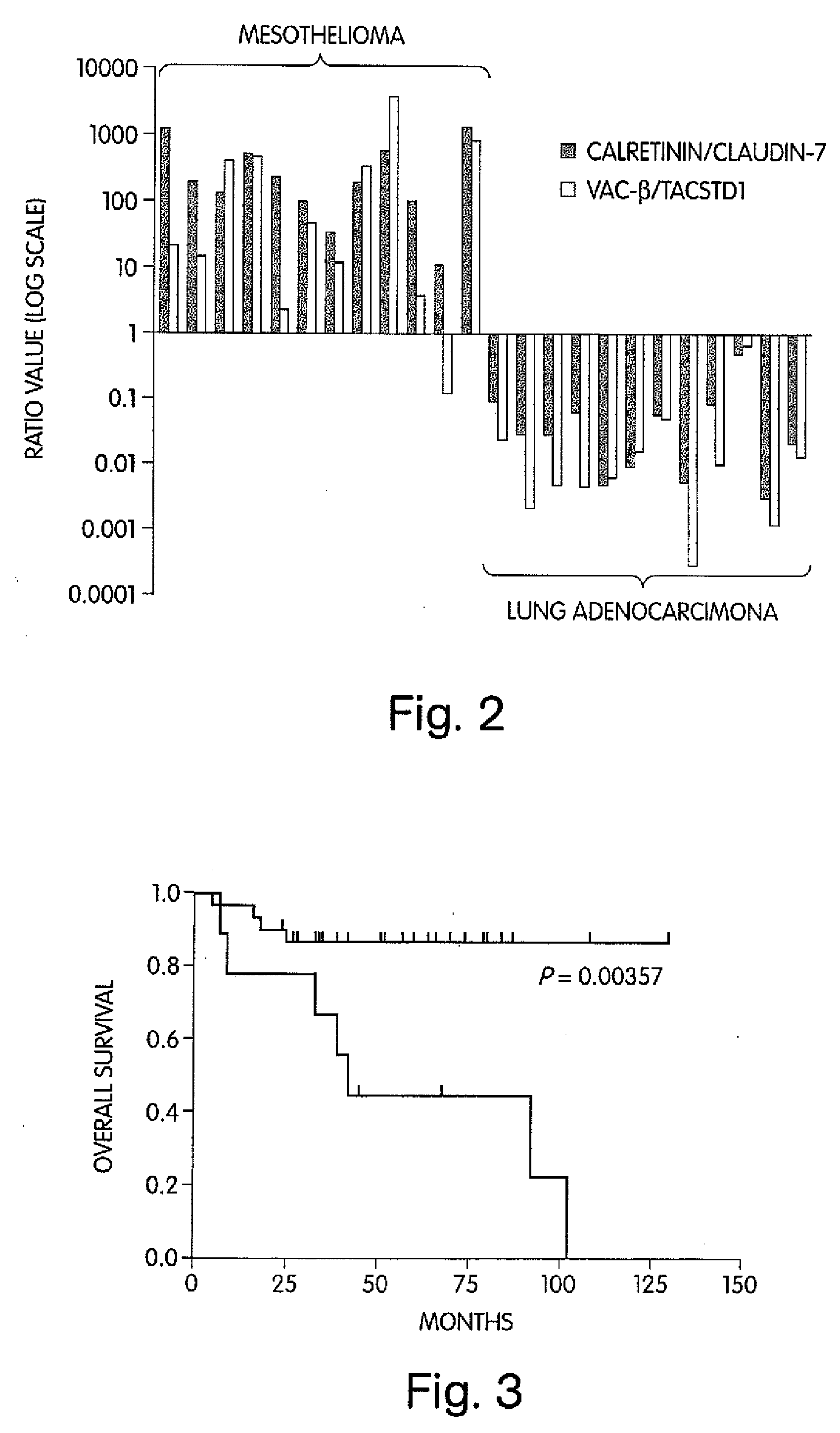
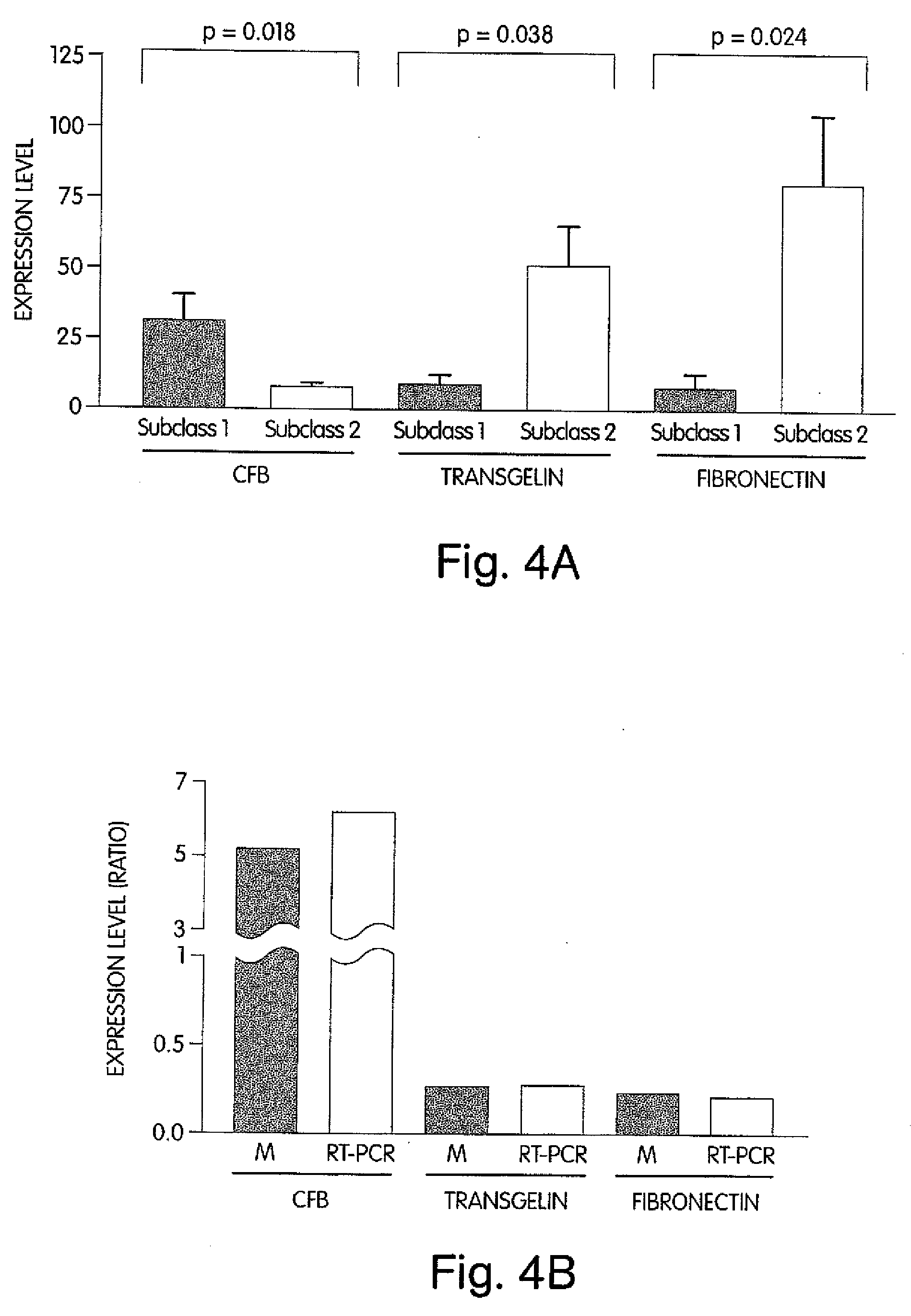
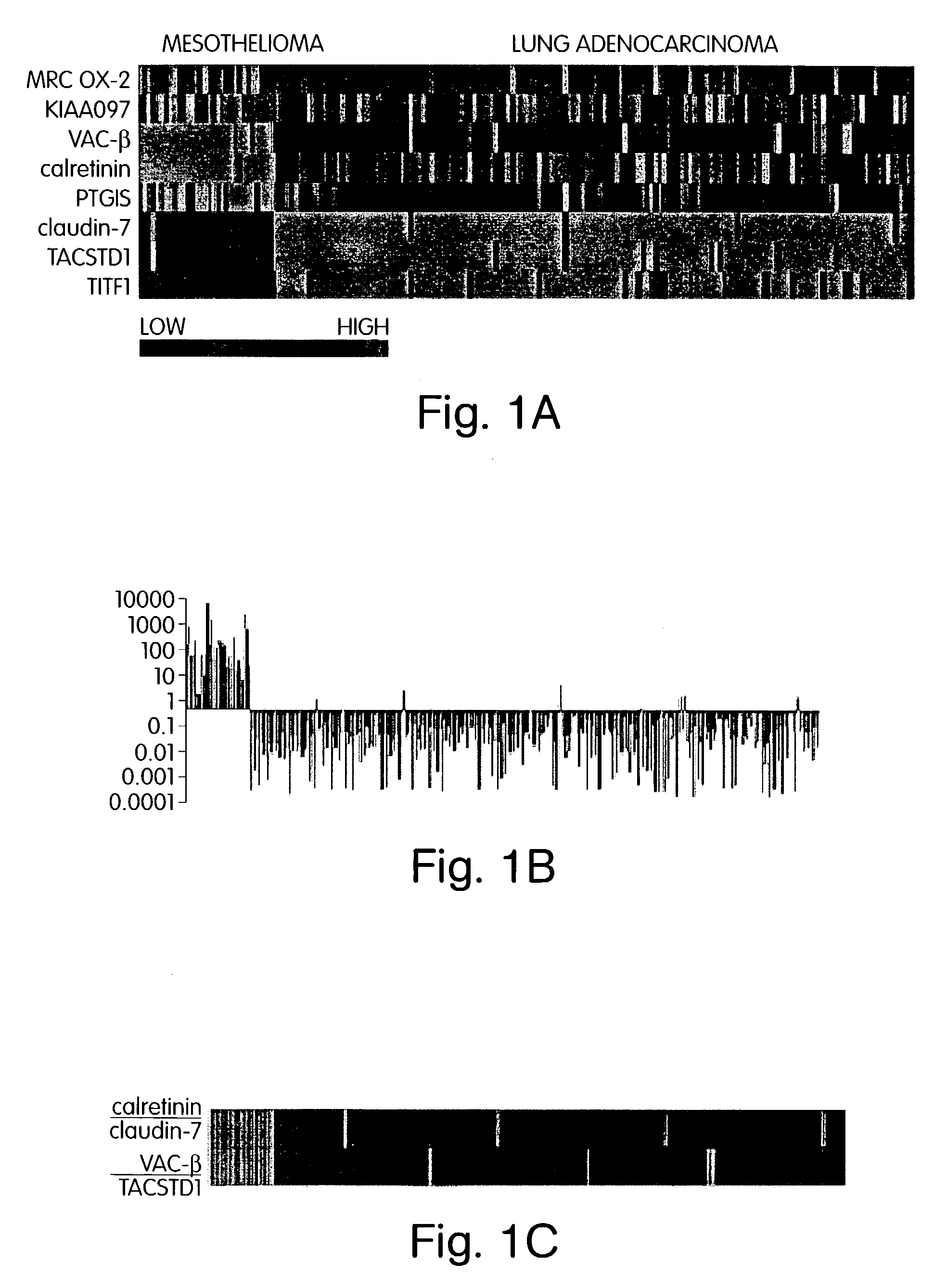
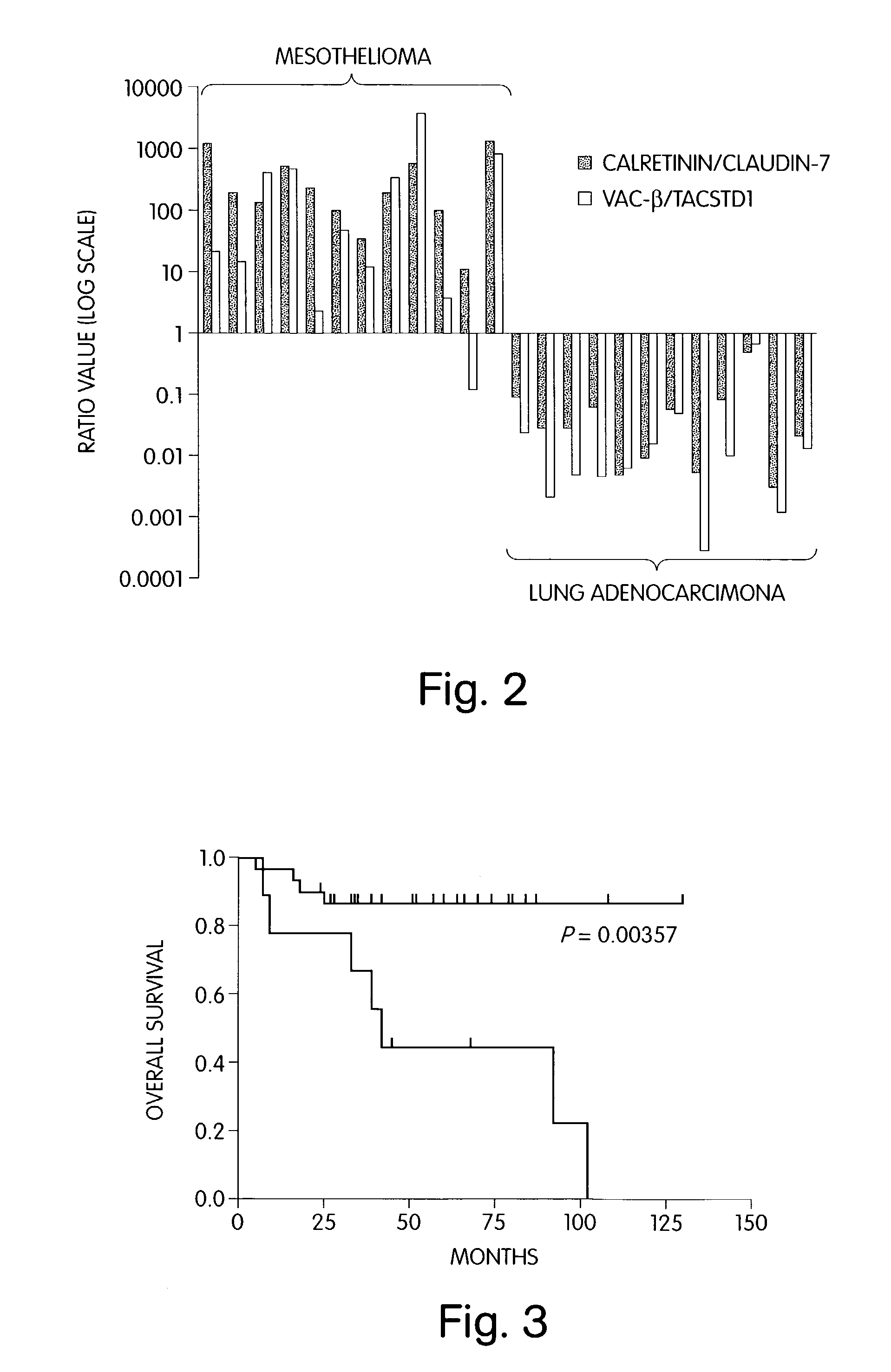
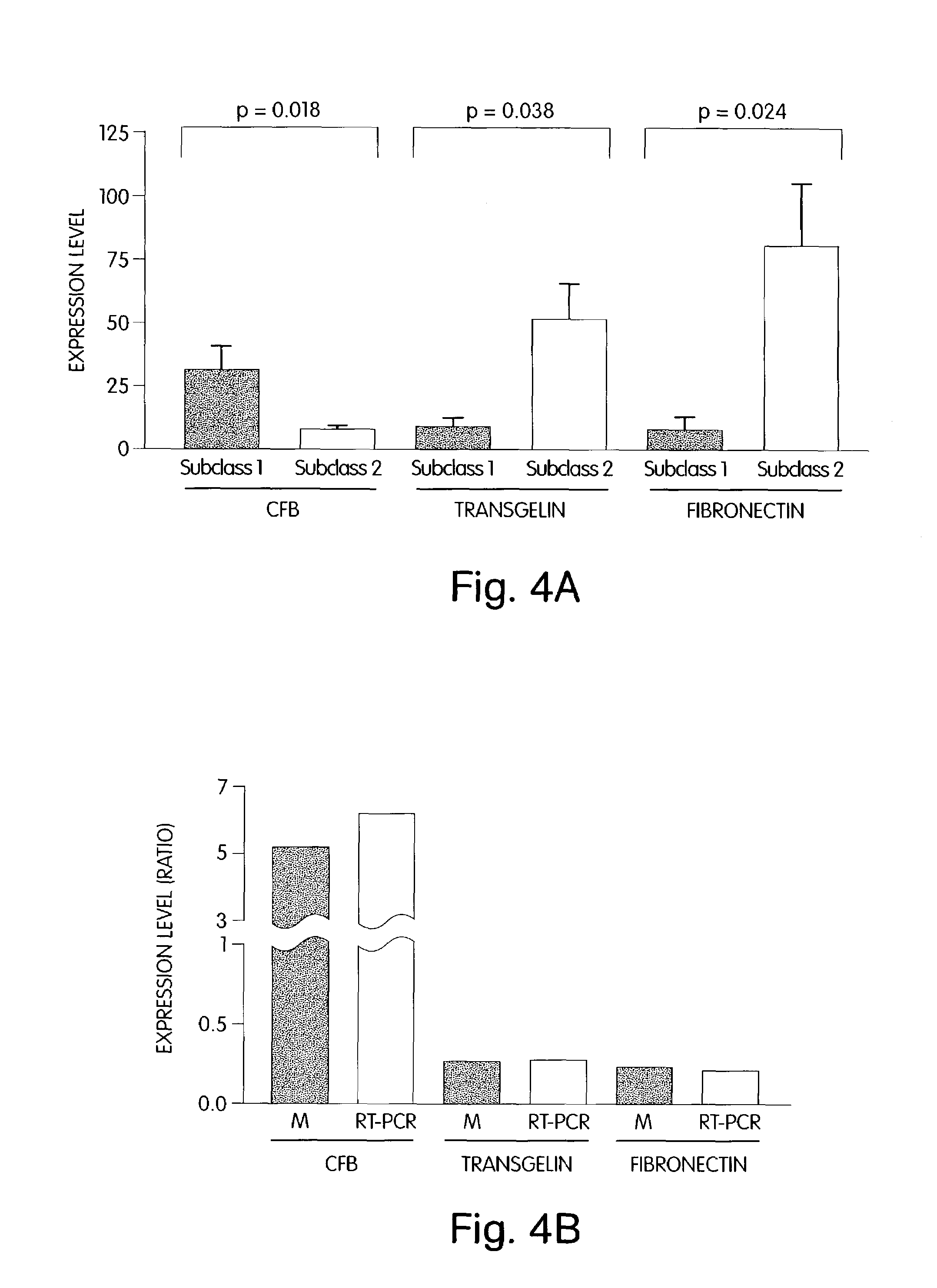
The invention provides methods for diagnosing biological states or conditions based on ratios of gene expression data from tissue samples, such as cancer tissue samples. The invention also provides sets of genes that are expressed differentially in malignant pleural mesothelioma. These sets of genes can be used to discriminate between normal and malignant tissues, and between classes of malignant tissues. Accordingly, diagnostic assays for classification of tumors, prediction of tumor outcome, selecting and monitoring treatment regimens and monitoring tumor progression / regression also are provided.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +1
Diagnostic and prognostic tests
InactiveUS7622260B2Simple powerEasy to adaptMicrobiological testing/measurementBiological testingTissue sampleExpression gene
The invention provides methods for diagnosing biological states or conditions based on ratios of gene expression data from tissue samples, such as cancer tissue samples. The invention also provides sets of genes that are expressed differentially in malignant pleural mesothelioma. These sets of genes can be used to discriminate between normal and malignant tissues, and between classes of malignant tissues. Accordingly, diagnostic assays for classification of tumors, prediction of tumor outcome, selecting and monitoring treatment regimens and monitoring tumor progression / regression also are provided.
Owner:WESLEYAN UNIVERSITY +1
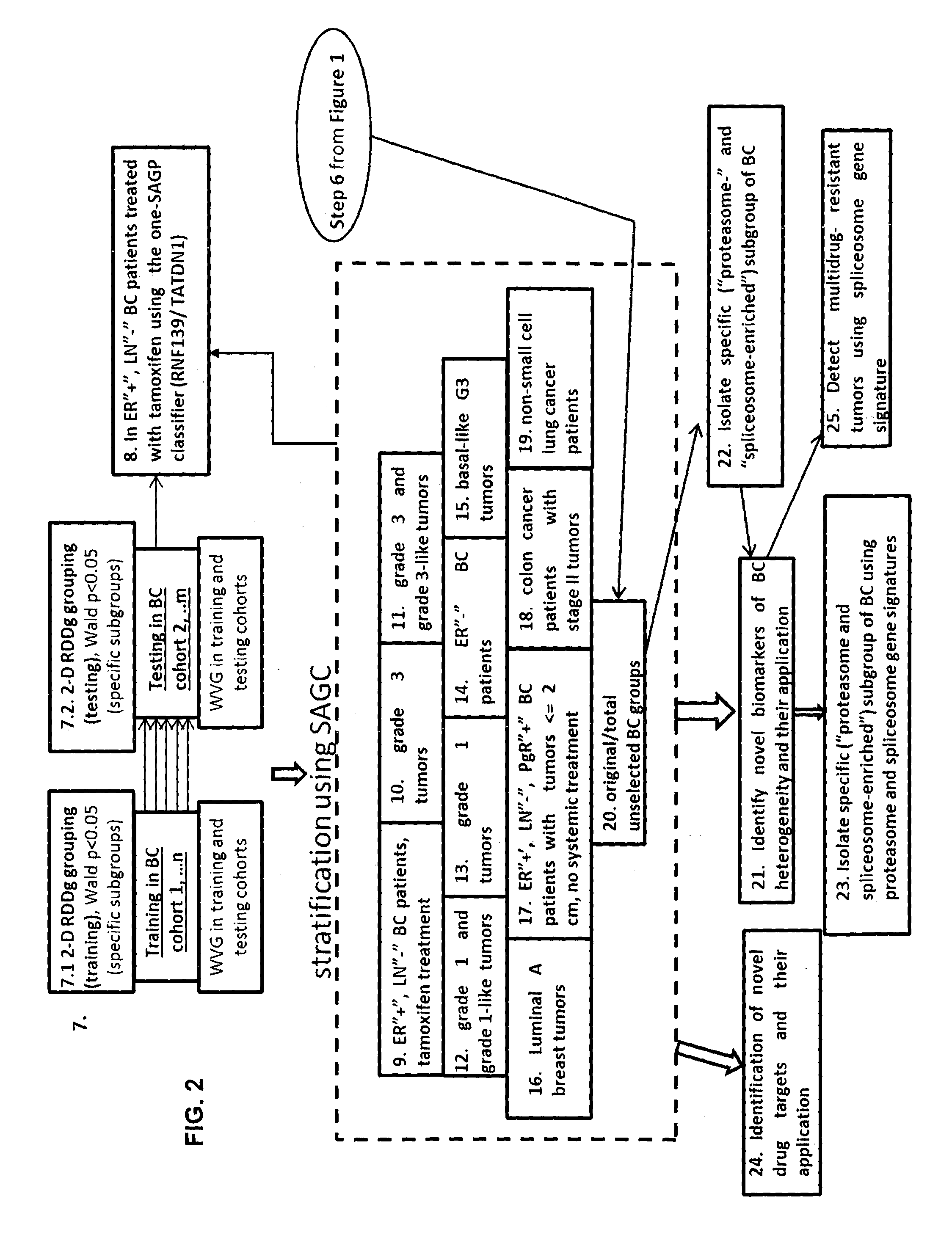
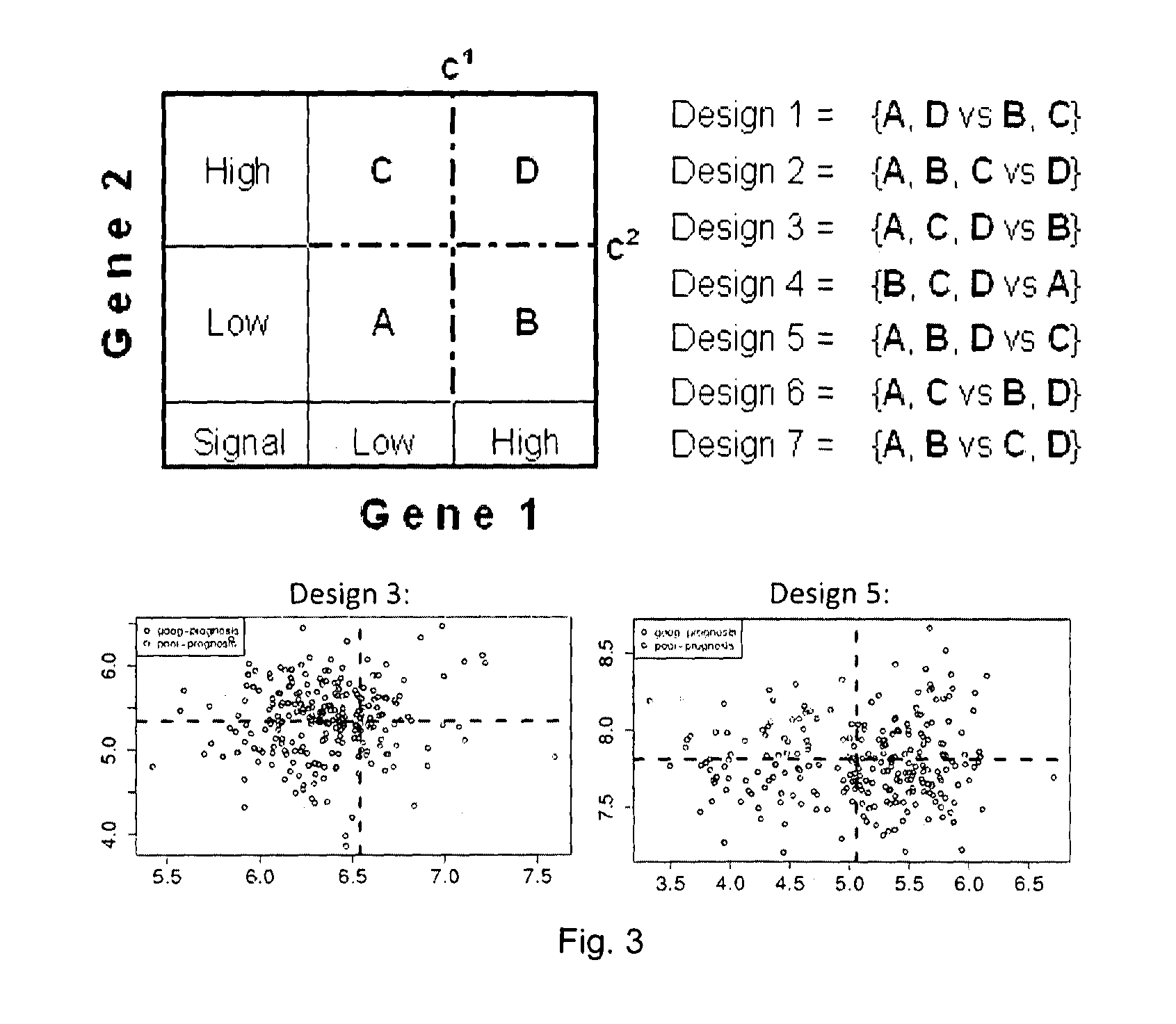
Sense-antisense gene pairs for patient stratification, prognosis, and therapeutic biomarkers identification
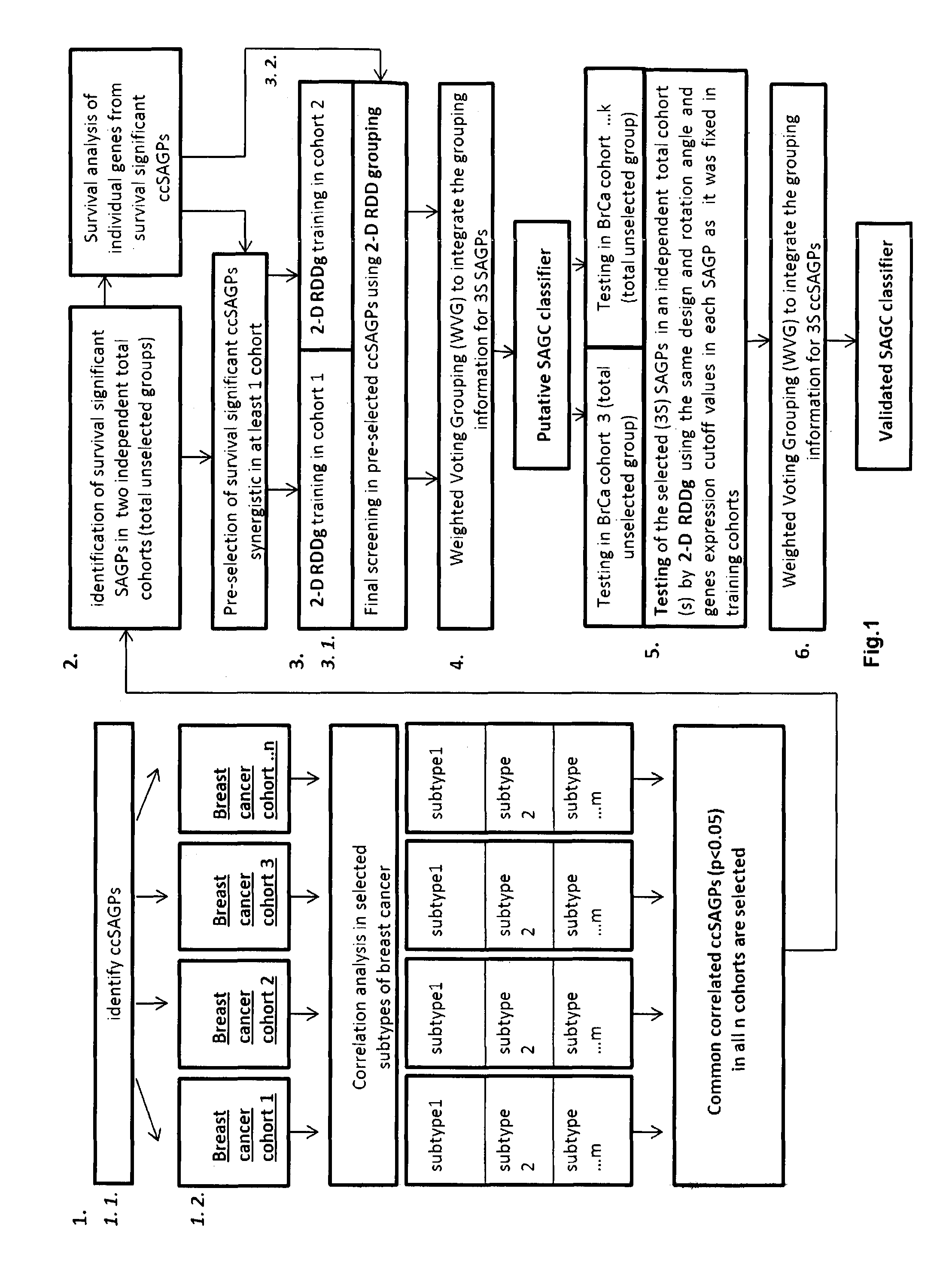
InactiveUS20160259883A1Quality improvementHighly prognostically significantMicrobiological testing/measurementLibrary screeningPrognostic signaturePatient stratification
The present invention relates to a method of identification of clinically and genetically distinct sub-groups of patients subject to a medical condition, particularly breast, lung, and colon cancer patients using a composition of respective gene expression values for certain gene pairs. Sense-antisense gene pairs (SAGPs) which are relevant for a medical condition and the disease prognosis are used by the method to generate statistical models based on the expression values of the SAGPs. SAGPs for which the statistical models are found to have high value in prognosis of the variation of medical condition and the diseases are selected and integrated in the prognostic signature including specified parameters (e.g. cut-off values) of the prognostic model. It further relates to using respective gene expression values for these genes to predict patient′ risk groups (in context of patient's survival or / and disease progression) and to using the predicted groups for identification of patient risk, and specific and robust prognostic biomarkers with mechanistic interpretations of biological changes (associated with the gene signatures) appropriating for an implementation of therapeutic targeting.
Owner:AGENCY FOR SCI TECH & RES
Diagnostic and prognostic methods for lung disorders using gene expression profiles from nose epithelial cells
InactiveUS20120190567A1Less invasiveDiagnosis, prognosis, and follow-upMicrobiological testing/measurementLibrary screeningLess invasiveLung cancer
The present invention provides methods for diagnosis and prognosis of lung cancer using expression analysis of one or more groups of genes, and a combination of expression analysis from a nasal epithelial cell sample. The methods of the invention provide far less invasive method with a superior detection accuracy for lung cancer when compared to any other currently available method for lung cancer diagnostic or prognosis. The invention also provides methods of diagnosis and prognosis of other lung diseases, such as lung cancer.
Owner:TRUSTEES OF BOSTON UNIV
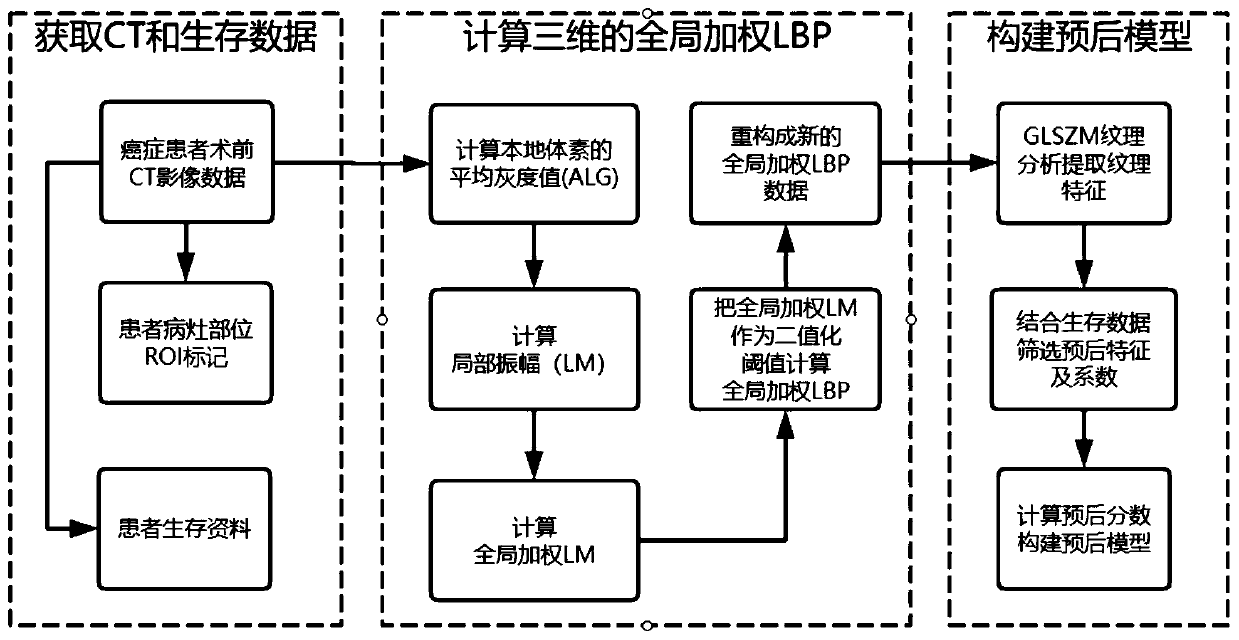
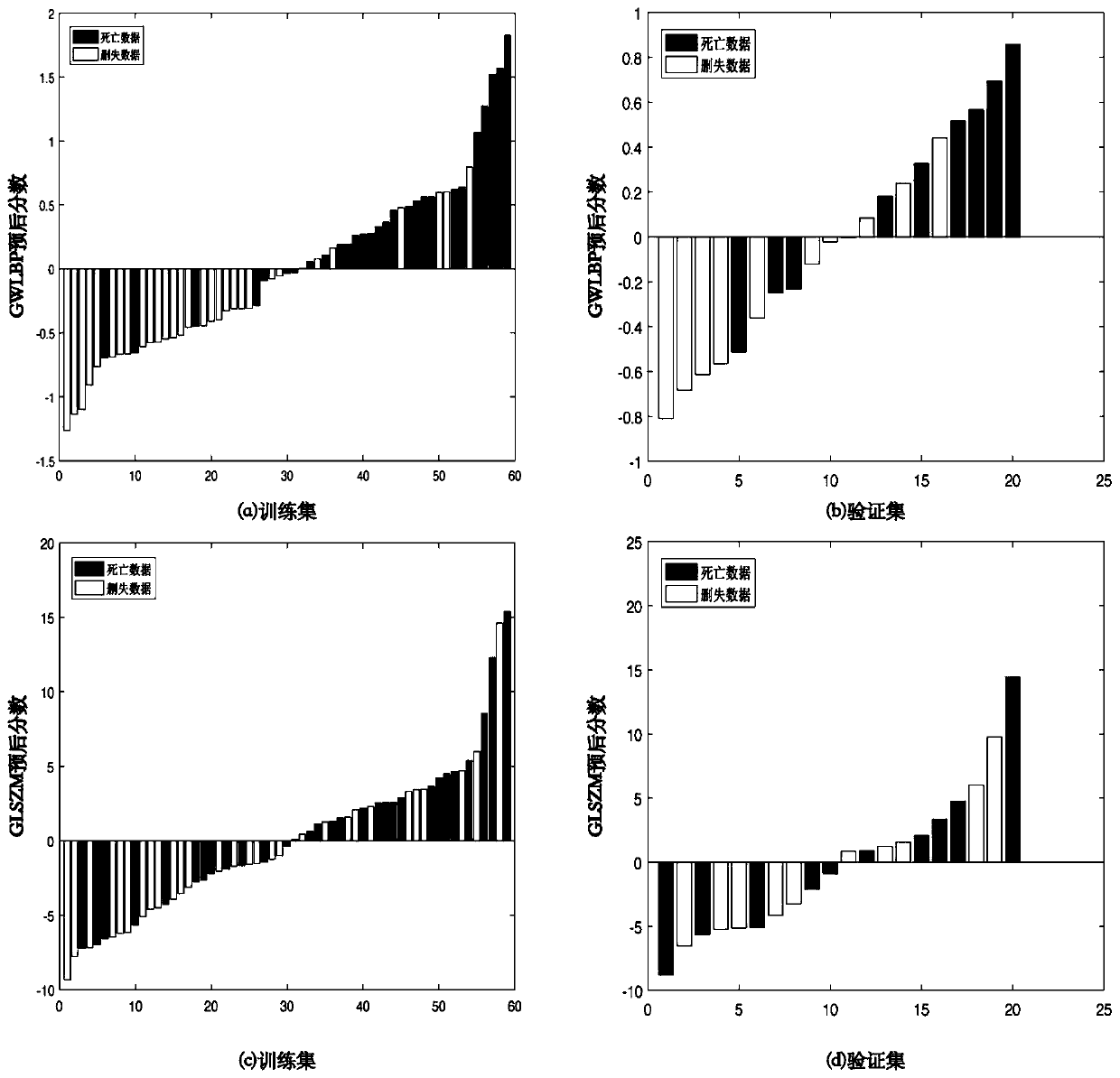
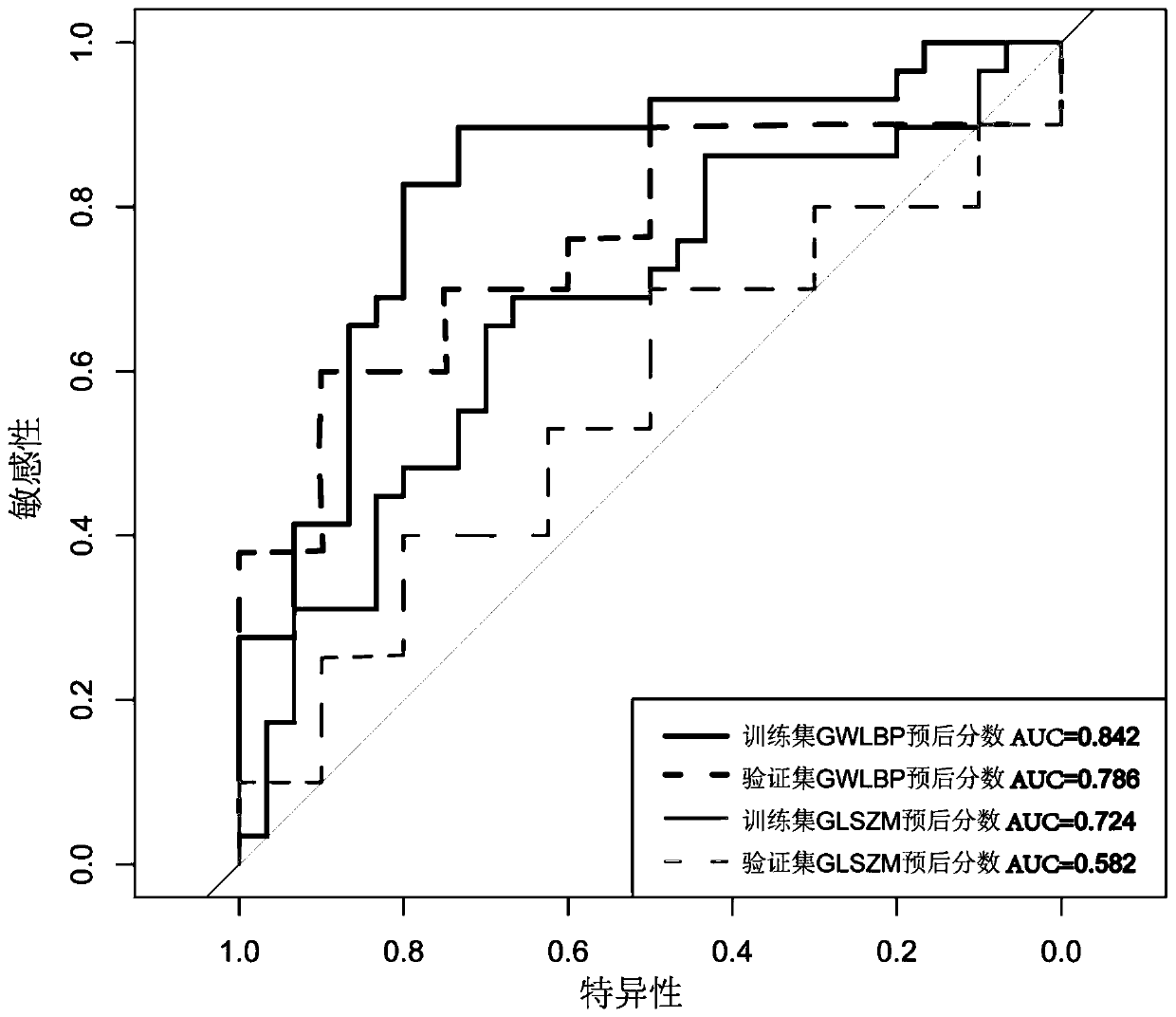
Cancer prognosis model construction method combining global weighted LBP and texture analysis
ActiveCN111340770AHighlight tumor featuresThe build result is validMedical simulationImage enhancementComputed tomographyImaging data
The invention discloses a cancer prognosis model construction method combining global weighted LBP (Local Binary Pattern) and texture analysis. The cancer prognosis model construction method comprisesthe following steps: acquiring original preoperative CT (Computed Tomography) image data of a cancer patient, marks of focus parts and survival data; calculating a three-dimensional global weighted LBP for the original CT data, and reconstructing new image data, namely global weighted LBP data; for the obtained global weighted LBP data, using GLSZM texture analysis to extract texture features. Texture analysis characteristics extracted by the method provided by the invention highlight tumor characteristics of a patient, and a prognosis model construction result is more effective; GLSZM is used for representing texture features, the effects in the aspects of texture consistency, rotation invariance and aperiodicity are remarkable, and the method has better performance than a co-occurrencematrix and a travel matrix in the aspect of cell nucleus and CT image texture.
Owner:TAIYUAN UNIV OF TECH
Liver cancer prognosis related serum miRNA markers and application of detection kit thereof
ActiveCN105647923AHigh clinical application valueBreakthrough innovationMicrobiological testing/measurementDNA/RNA fragmentationIndividualized treatmentSerum mirna
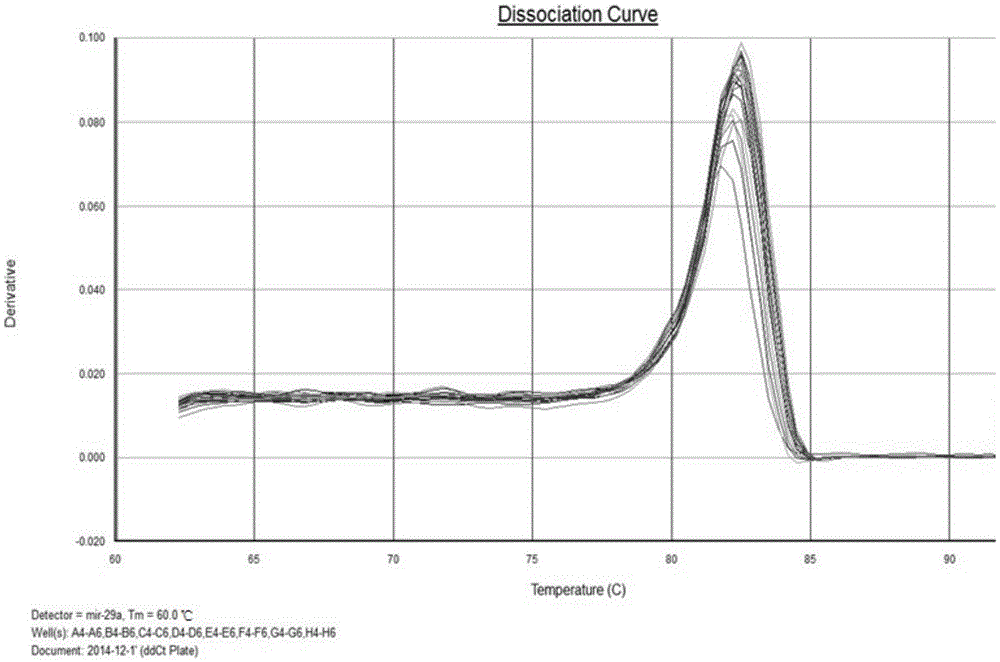
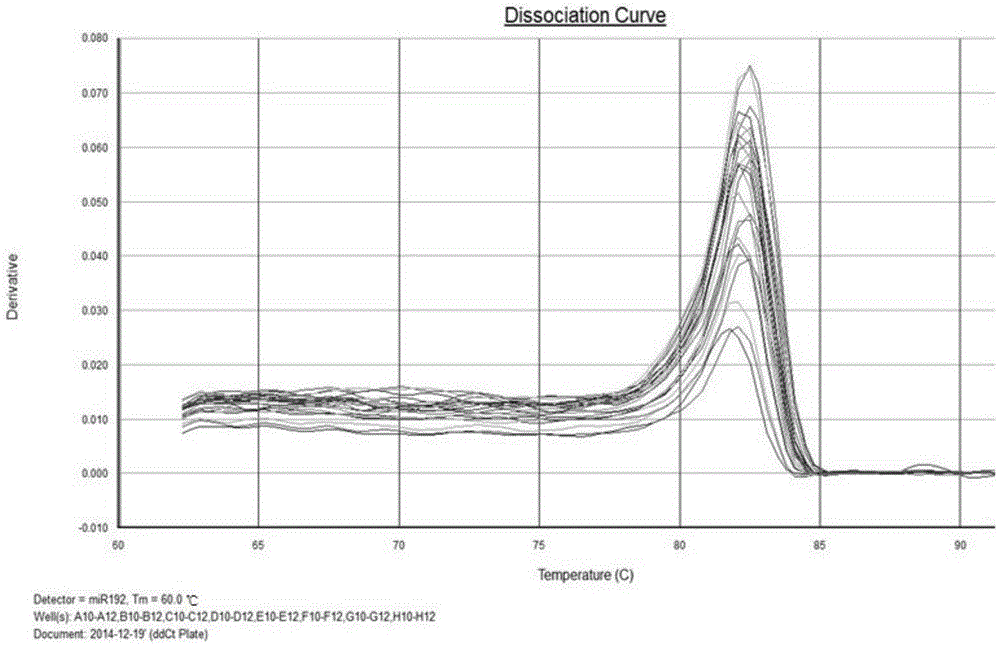
The invention discloses liver cancer prognosis related serum miRNA markers and an application of a detection kit thereof. The tumor markers are serum miR-29a-3p and miR-192-5p which are related with human liver cancer patients. According to preliminary work, liver cancer related serum miRNA expression profiles containing miR-29a-3p and miR-192-5p and having obvious expression difference are screened out through high-throughput deep sequencing, and qPCR (quantitative polymerase chain reaction) proves and finally confirms that miR-29a-3p and miR-192-5p are liver cancer risk (prognosis) related tumor markers. The markers and detection reagents can be used for preparing the rapid-detection pPCR kit, and the kit has the characteristics of rapidness and convenience in detection, high accuracy rate, no wound and the like; a prognostic model built through combination of miR-29a-3p, miR-192-5p and BCLC stage (Barcelona clinic liver cancer stage) can be applied to classification of risk degrees of the liver cancer patients, thereby guiding individualized treatment of the liver cancer patients.
Owner:SUN YAT SEN UNIV CANCER CENT
Gene expression markers for response to EGFR inhibitor drugs
InactiveUS20080318230A1Microbiological testing/measurementAntineoplastic agentsParaffin embeddedEGFR inhibitors
The present invention concerns prognostic markers associated with cancer. In particular, the invention concerns prognostic methods based on the molecular characterization of gene expression in paraffin-embedded, fixed samples of cancer tissue, which allow a physician to predict whether a patient is likely to respond well to treatment with an EGFR inhibitor.
Owner:GENOMIC HEALTH INC +1
Pulmonary embolism clinical risk and prognosis scoring method and system
PendingCN113327679AEasy to usePredictable risk of deathHealth-index calculationMedical automated diagnosisStatistical analysisMedical treatment
The invention provides a pulmonary embolism clinical risk and prognosis scoring method and a system, and the method comprises the steps: carrying out the screening and feature extraction of medical data, and obtaining influence factors corresponding to death; performing single-factor logistic regression analysis on the influence factors, determining variables of a prognosis model, and establishing a logistic regression model; obtaining a result OR value through statistical analysis; assigning each risk factor based on the OR value, and establishing a risk prediction scoring system of pulmonary embolism to predict the death risk of the pulmonary embolism patient within 30 days; constructing an ROC curve, and comparing the areas under the curve of the three scoring systems by using clinical data of the verification group, the modeling group and the modeling group + the verification group; and predicting a result according to the optimal truncation value. The risk and prognosis scoring method can evaluate the death risk of the pulmonary embolism patient within 30 days, and high-risk and low-risk patients can be identified for doctors to refer to and formulate a diagnosis scheme.
Owner:上海市闵行区中心医院
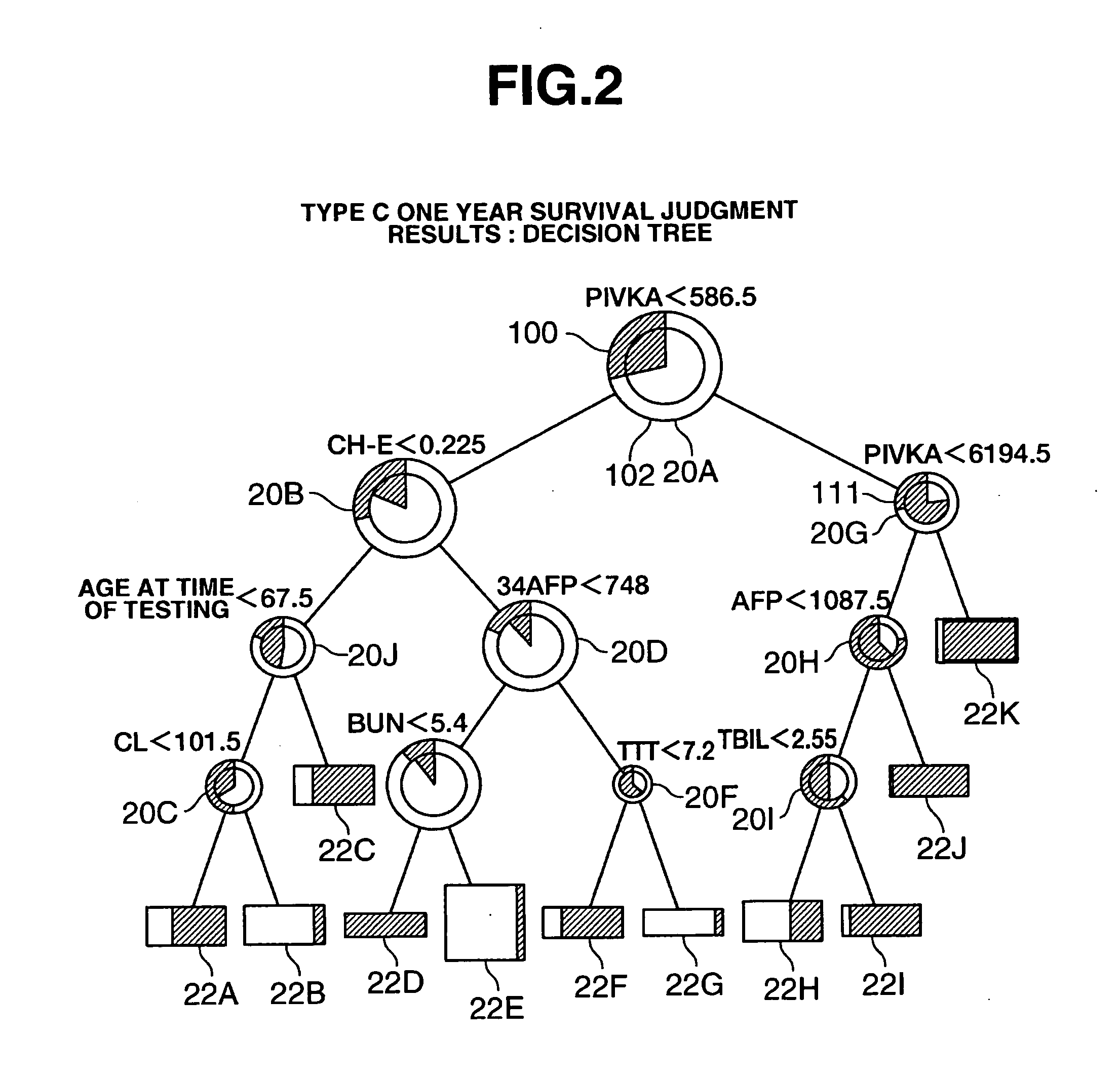
Method of Preparing Disease Prognosis Model, Disease Prognosis Prediction Method using this Model, Prognosis Prediction Device Based on this Model, and Program for Performing the Device and Storage Medium Wherein Said Program is Stored
InactiveUS20070208514A1Add supportAccurate analysisMedical simulationMedical data miningPrognosis predictionDisease cause
The present invention relates to a disease prognosis prediction modeling method for preparing a model for predicting the prognosis of a specified disease from clinical laboratory test values for the disease by means of a computer, the method comprising the steps of: inputting a plurality of actually measured clinical laboratory test values for the disease and actual measured values of the prognoses into the computer; processing these values by a data mining method to determine one or a plurality of clinical laboratory test items which have an influence on the prognosis of the disease; determining a priority of the items with respect to the prognosis in a case where there are a plurality of the items; and establishing a judgment routine in which correlation of the plurality of clinical laboratory test items and the clinical laboratory test value ranges of the test items with the predicted value of the prognosis is stipulated on the basis of the priority, wherein the judgment routine is used as the model.
Owner:EISIA R&D MANAGEMENT CO LTD
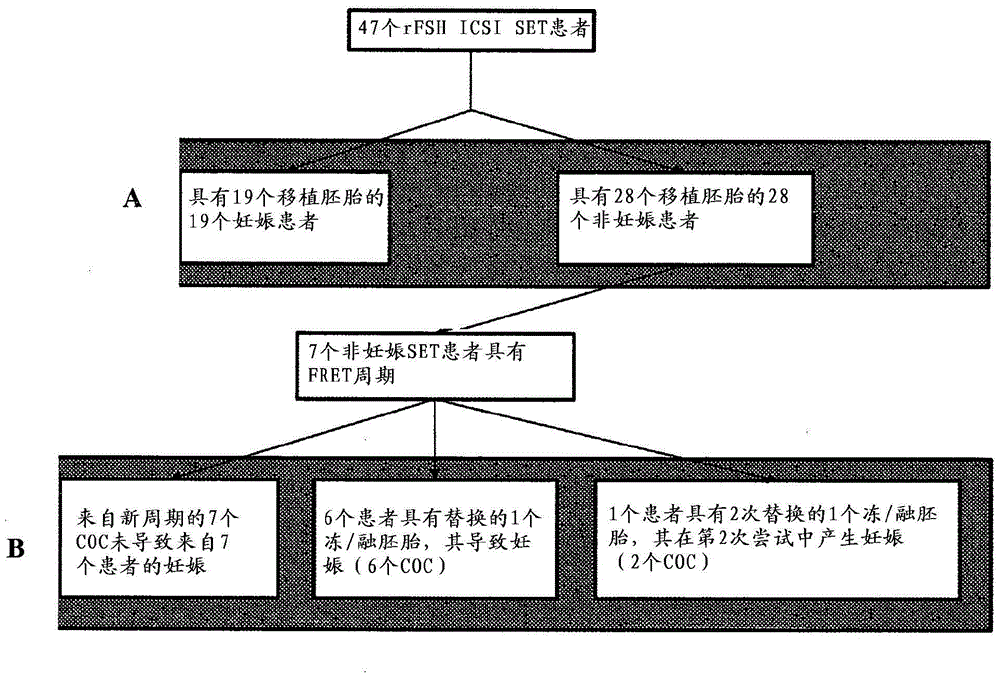
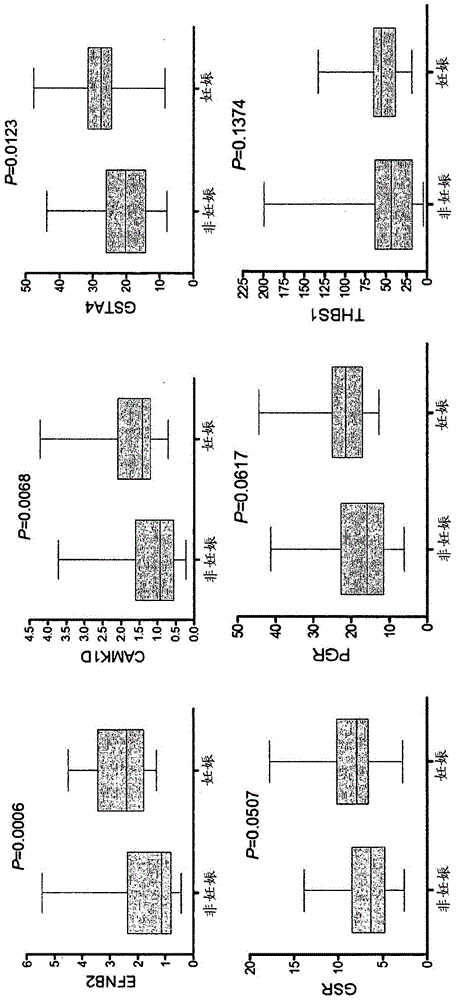
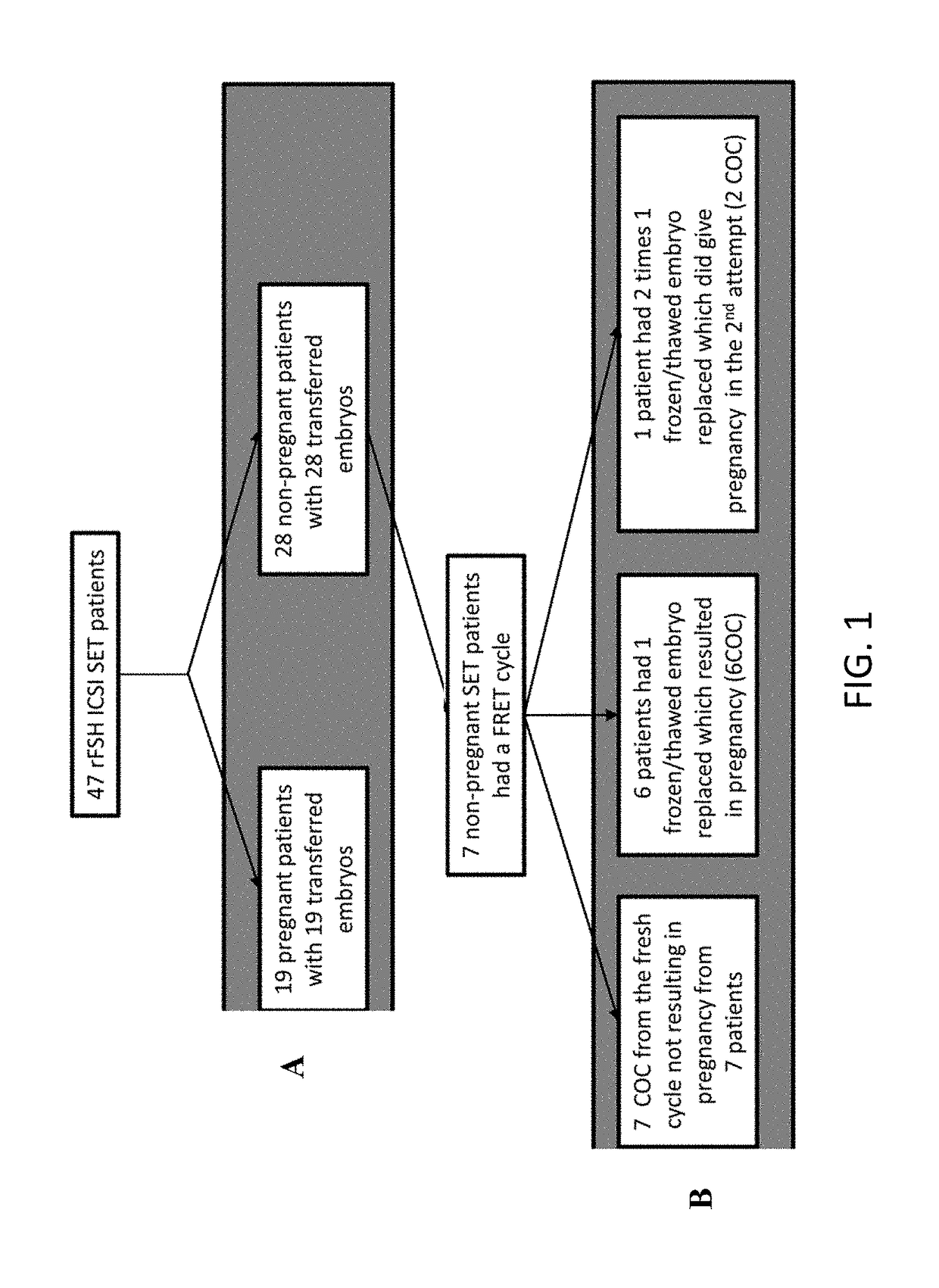
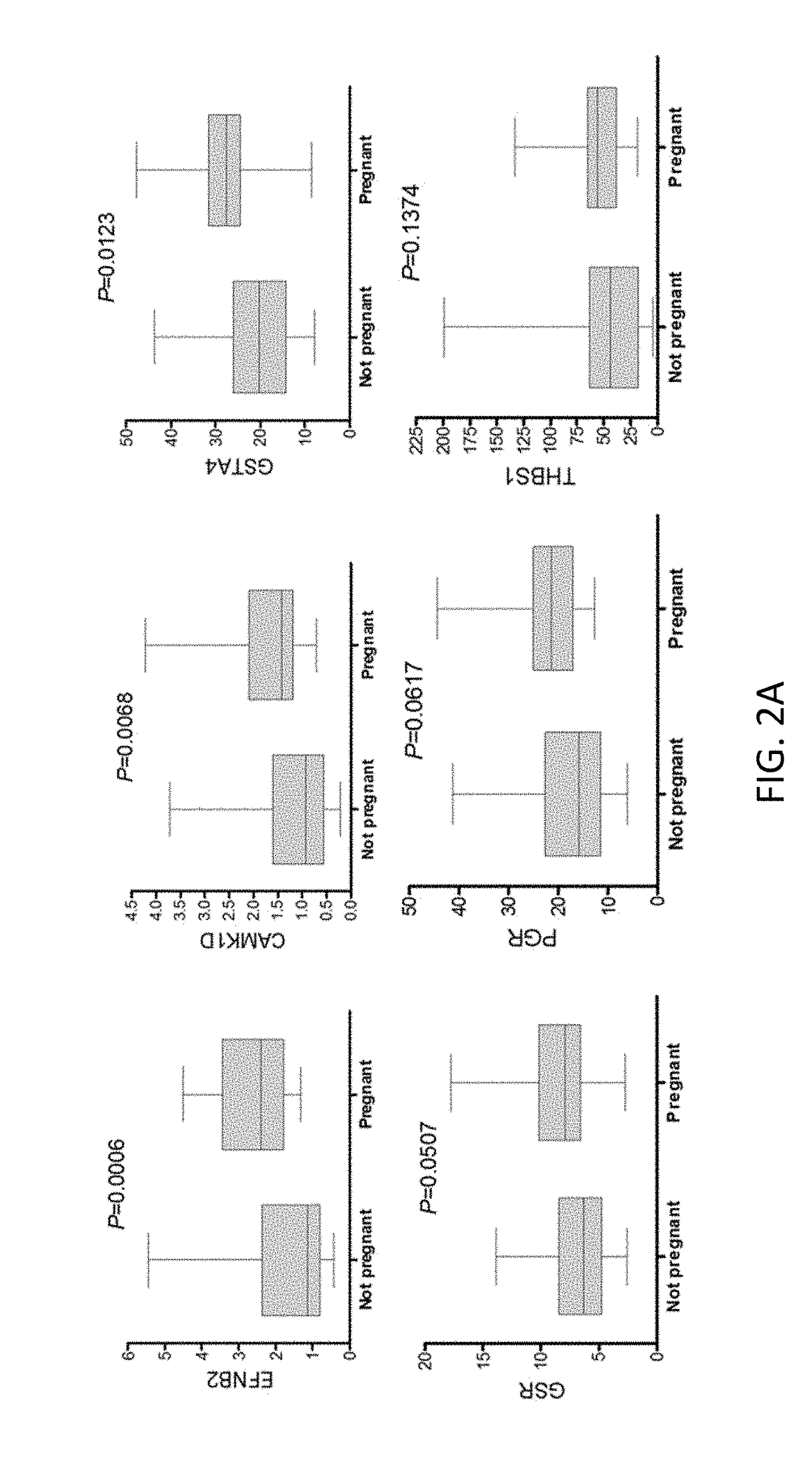
Marker genes for oocyte competence
Cumulus cell (CC) gene expression is explored as an additional method to morphological scoring to choose the embryo with the highest chance to pregnancy. The present invention relates to a novel method of identifying biomarker genes for evaluating the competence of a mammalian oocyte in giving rise to a viable pregnancy after fertilization, based on the use of live birth and embryonic development as endpoint criteria for the oocytes to be used in an exon level analysis of potential biomarker genes. The invention further provides CC-expressed biomarker genes thus identified, as well as prognostic models based on the biomarker genes identified using the methods of the present invention.
Owner:VRIJE UNIV BRUSSEL
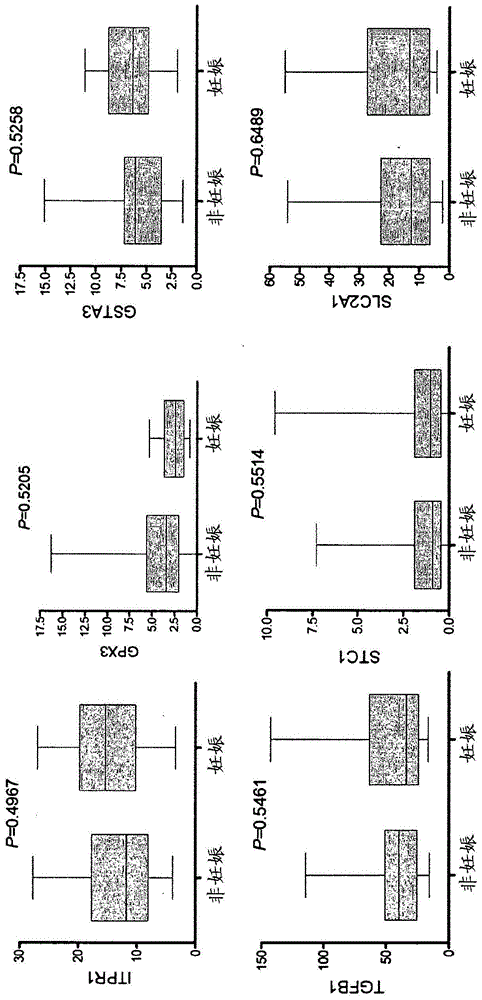
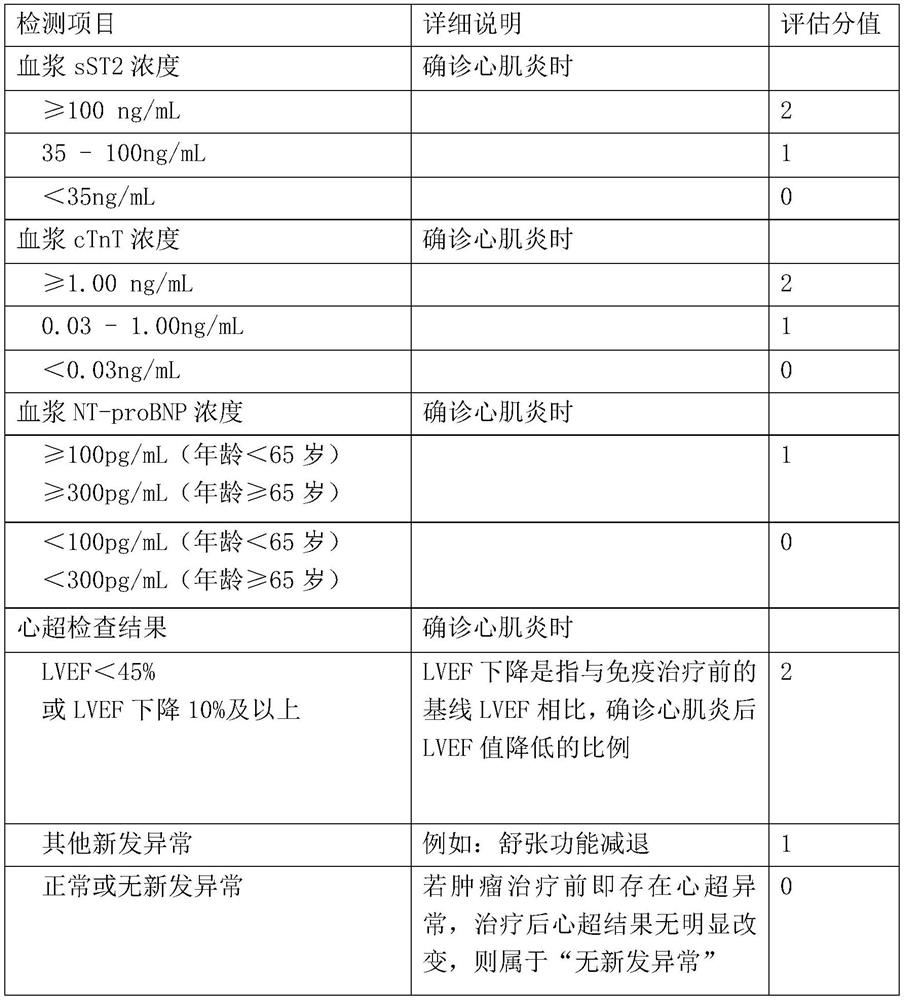
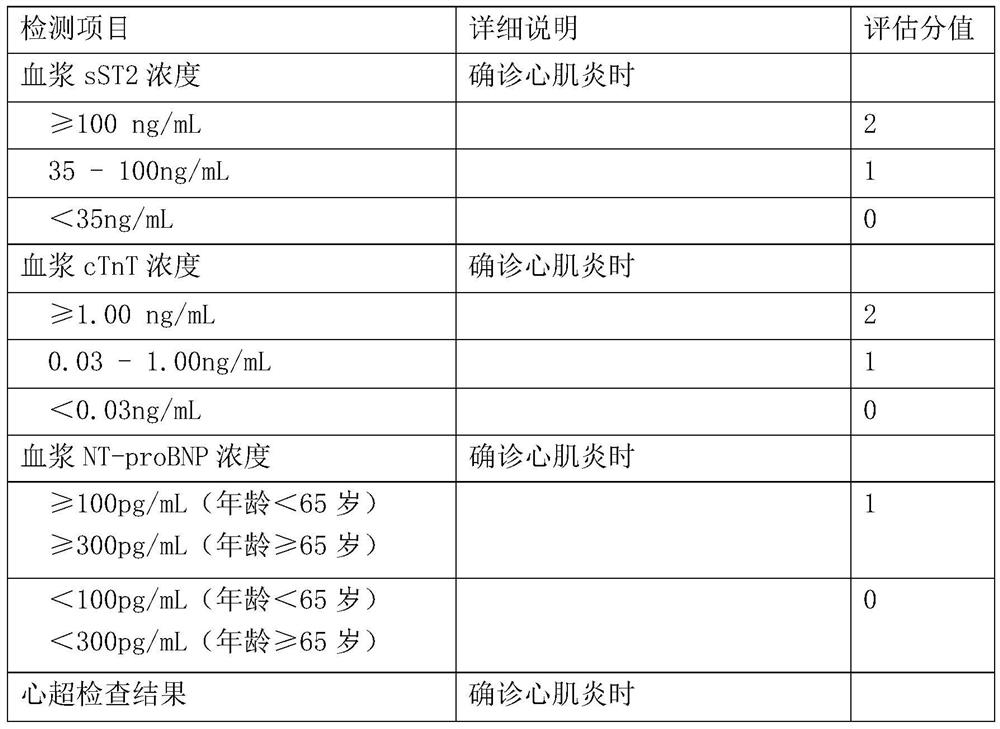
Prognosis model for evaluating tumor immunotherapy related myocarditis based on sST2 and application thereof
PendingCN111863259AEffectively formulateReasonable treatment planHealth-index calculationMedical automated diagnosisMyocarditisCardiac functioning
The invention relates to a prognosis model for evaluating tumor immunotherapy related myocarditis based on sST2 and application thereof, and belongs to the technical field of biomedical detection. Theinvention provides a model for evaluating prognosis of tumor immunotherapy related myocarditis based on sST2. Immune myocarditis is a few but severe adverse reaction in tumor immunotherapy, and sST2is a novel heart marker and participates in the process of cardiac dysfunction caused by cardiac hypertrophy, myocardial fibrosis, ventricular remodeling and the like. The invention provides an sST2-based model for evaluating tumor immunotherapy related myocarditis prognosis, which is beneficial to providing reference information for disease prognosis of immunotherapy related myocarditis patientsso as to assist in clinically formulating a reasonable diagnosis and treatment scheme.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV
Bladder cancer pathomics intelligent diagnosis method based on machine learning and prognosis model thereof
PendingCN112435743AAlleviate shortagesEffective automatic pathological diagnosisMedical simulationMedical data miningMicroscopic imageBladder cancer patient
The invention relates to a bladder cancer pathomics intelligent diagnosis method based on machine learning and a prognosis model thereof, and the method is characterized in that the method comprises the following steps: S1, preforming data acquisition; S2, processing a microscopic pathology image; S3, performing bladder cancer pathological image feature extraction; S4, constructing and inspectinga bladder cancer automatic pathological diagnosis model based on machine learning; and S5, constructing and checking a bladder cancer survival prognosis prediction model based on machine learning. Thebladder cancer pathomics intelligent diagnosis method has the advantages that the bladder cancer pathomics intelligent diagnosis method is constructed based on pathological section microscopic imagemachine learning, effective automatic pathological diagnosis can be achieved, pathological diagnosis is expected to be further promoted to be developed to the efficient and accurate field, and the current situation that domestic pathology physicians are in shortage is relieved; the postoperative survival condition of the bladder cancer patient can be efficiently and accurately predicted, and important guidance opinions are provided for clinical decisions of clinicians.
Owner:SHANGHAI FIRST PEOPLES HOSPITAL
Evaluation model device for prognosis of esophageal squamous cell carcinoma and modeling method thereof
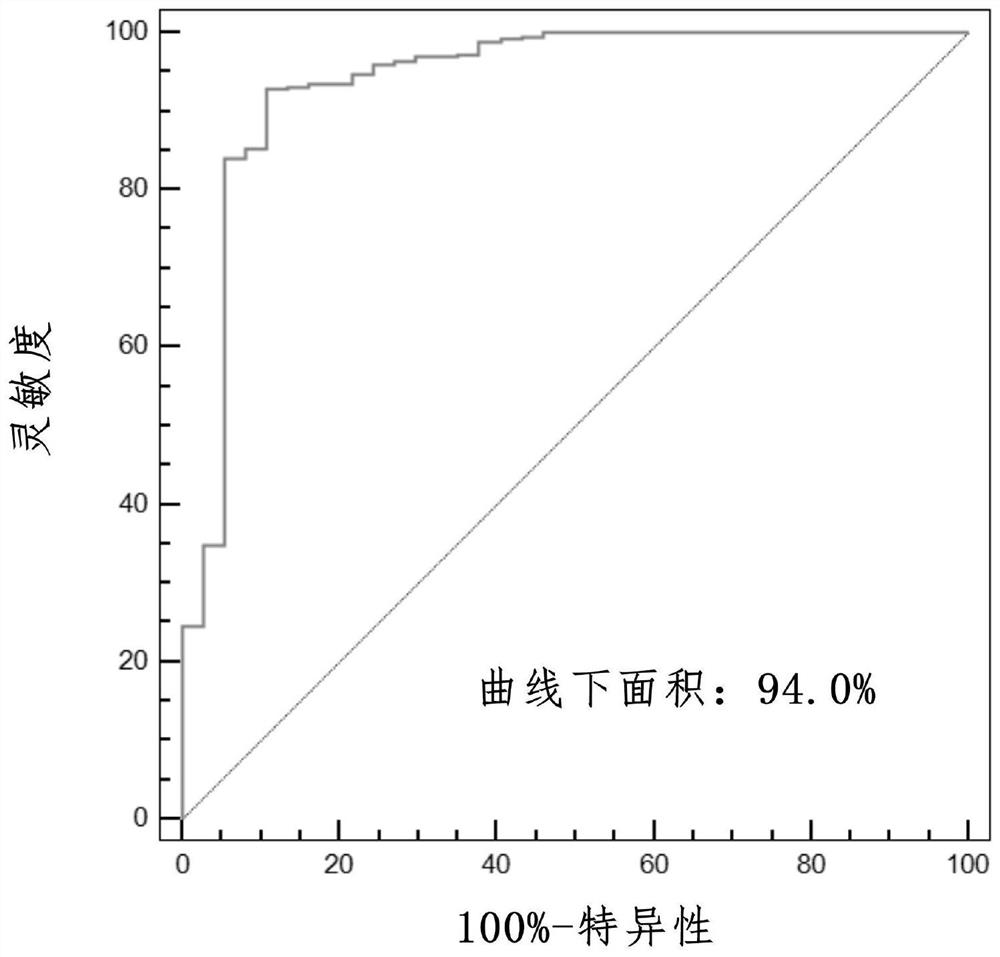
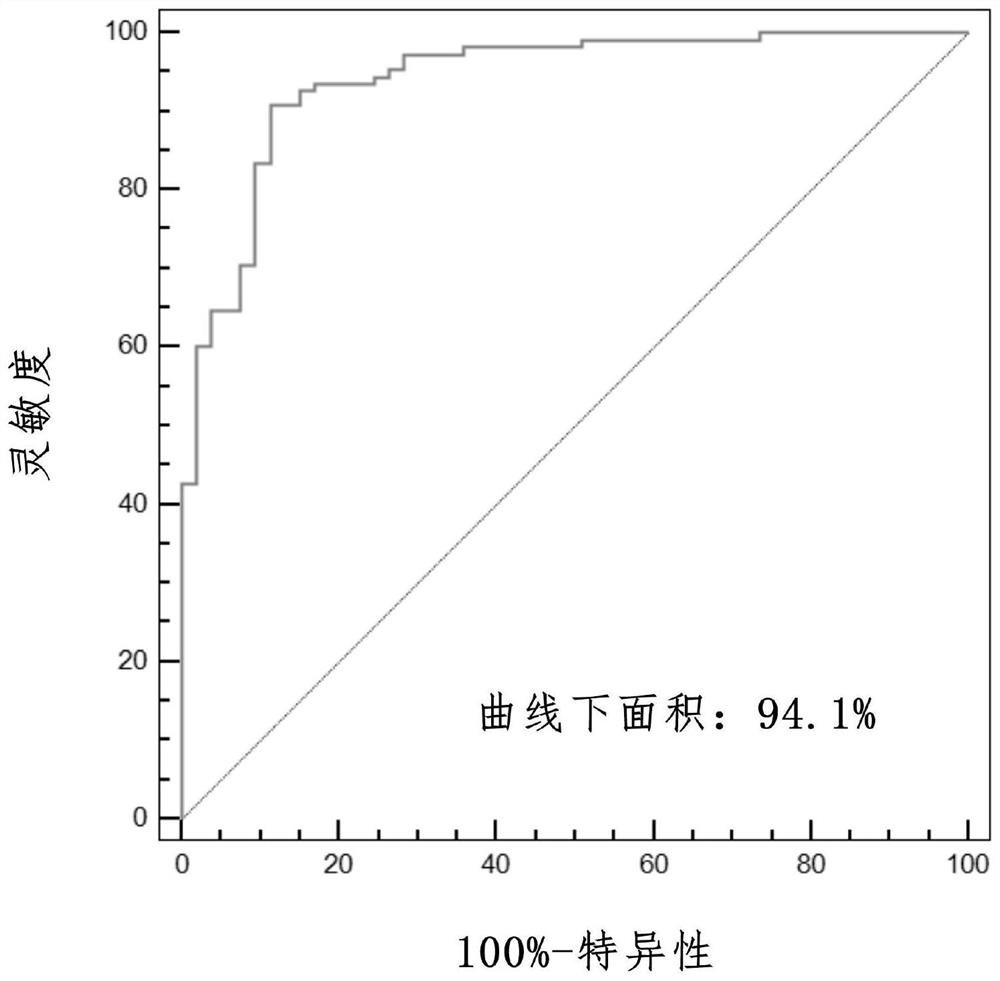
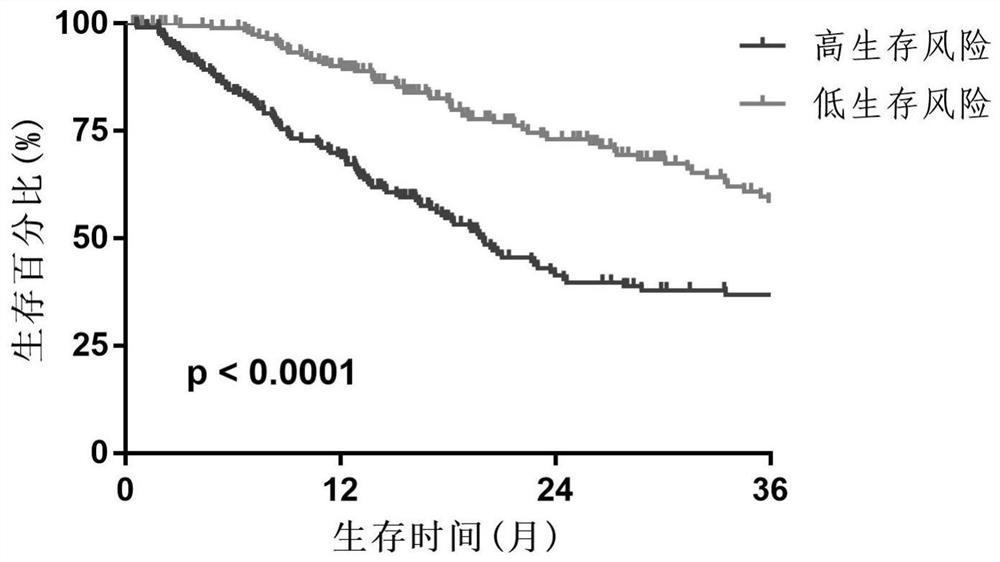
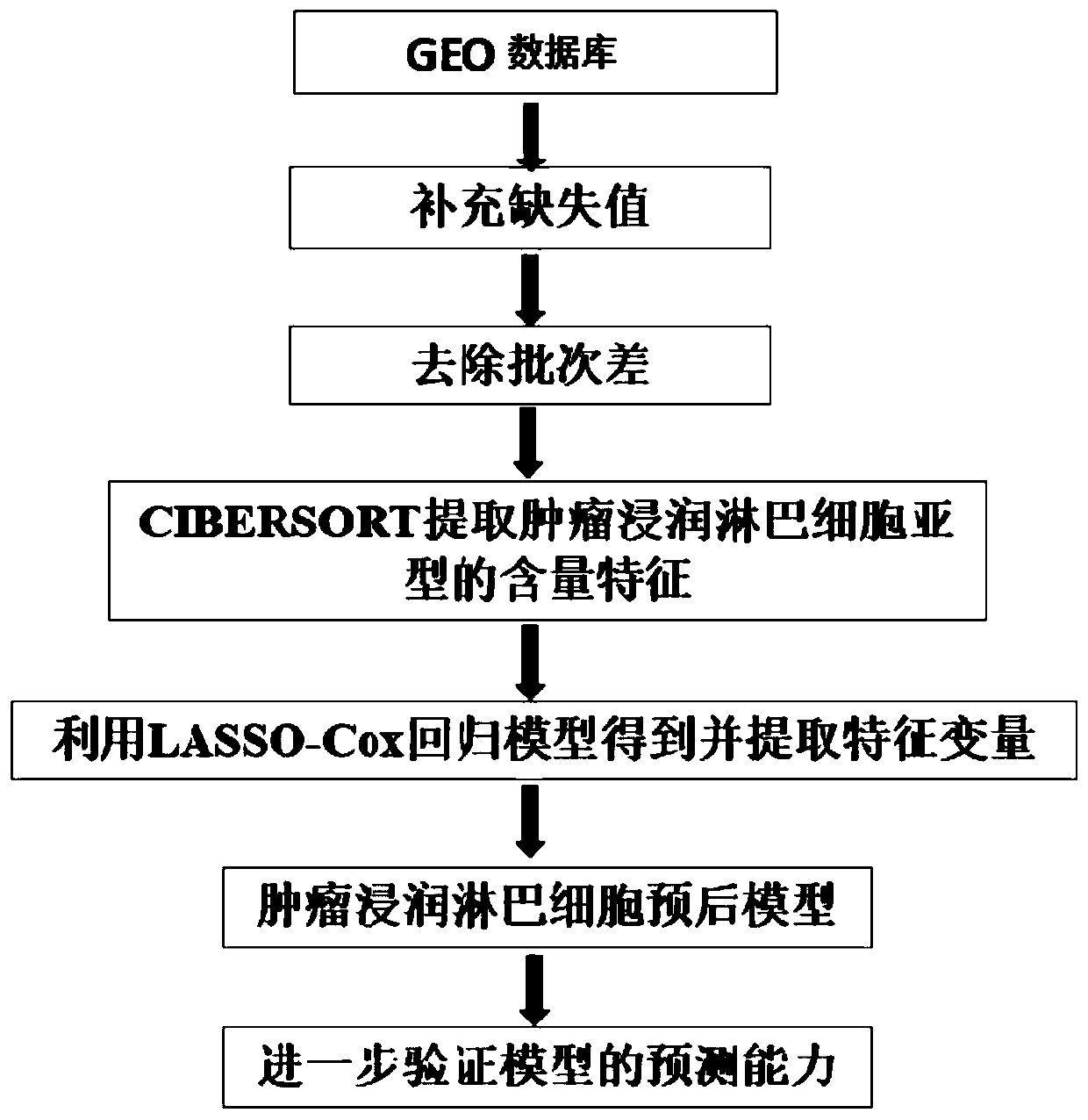
PendingCN111524597AMedical data miningHealth-index calculationData acquisitionData acquisition module
The invention discloses an evaluation model device for prognosis of esophageal squamous cell carcinoma and a modeling method thereof. The evaluation model device for prognosis of esophageal squamous cell carcinoma comprises a GEO database module, a data acquisition module, a data preprocessing module, a content feature extraction module, a feature variable extraction module and a prognosis model establishment module, wherein the data acquisition module is used for acquiring gene expression data and prognosis data related to esophageal squamous cell carcinoma in the GEO database module; the data preprocessing module is used for preprocessing the gene expression data and the prognosis data related to esophageal squamous cell carcinoma; and the content feature extraction module is used for extracting content features of the data processed by the data preprocessing module. According to the invention, a tumor infiltration lymphocyte subtype related to survival time can be screened, and a prognosis model is established; and when new feature data exists, the survival time probability of a patient with esophageal squamous cell carcinoma can be obtained only by inputting the values of the feature data into a regression model, so the method has important significance in prognosis of esophageal squamous cell carcinoma.
Owner:FIRST HOSPITAL OF QINHUANGDAO
Application of model constructed based on PCD related gene combination in preparation of product for predicting prognosis of colonic adenocarcinoma
PendingCN114540499AGood prognostic predictive powerGood clinical prognosis predictive abilityMicrobiological testing/measurementBiostatisticsDrug targetChemo therapy
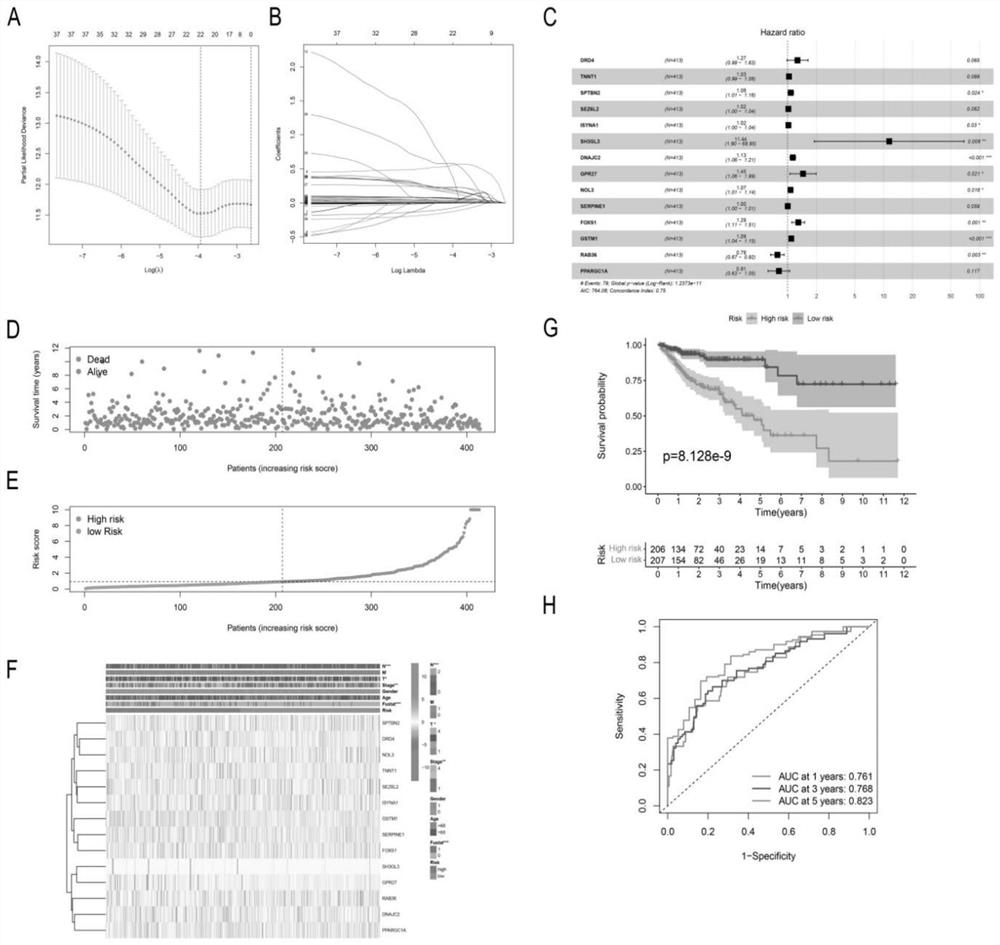
The invention discloses application of a model constructed based on a PCD related gene combination in preparation of a product for predicting prognosis of colonic adenocarcinoma. 14 PCD key genes differentially expressed in colorectal cancer, including (SPTBN2, DNAJC2, DRD4, SH3GL3, FOXS1, GSTM1, ISSYNA1, NOL3, GPR27, PPARGC1A, RAB36, SEZ6L2, SERPINE1 and TNNT1), are adopted as detection targets to construct a prognosis model PCDSig, the model has good clinical prognosis prediction ability for one year, two years and three years, the model can better distinguish high-risk people from low-risk people, and high-risk and low-risk patients have the characteristics of immune infiltration, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency, immune deficiency and the like. The response of immunotherapy is different from the aspects of drug target gene expression and chemotherapy drug sensitivity, target benefit people can be screened through the model, clinical precise and personalized treatment is effectively guided, and excessive treatment and patient condition delay are avoided. On the basis of the model, a column diagram for predicting the survival probabilities of the patient in one year, three years and five years is developed, so that the model is visualized, and conditions are provided for clinicians to judge the prognosis of the COAD patient.
Owner:郑州源创基因科技有限公司 +1
Immune-clinical feature combined prediction model for evaluating prognosis of lung adenocarcinoma
ActiveCN113215254AFacilitate personalized managementImproved prognostic accuracyMicrobiological testing/measurementBiostatisticsClinical prognosisPulmonary adenocarcinoma
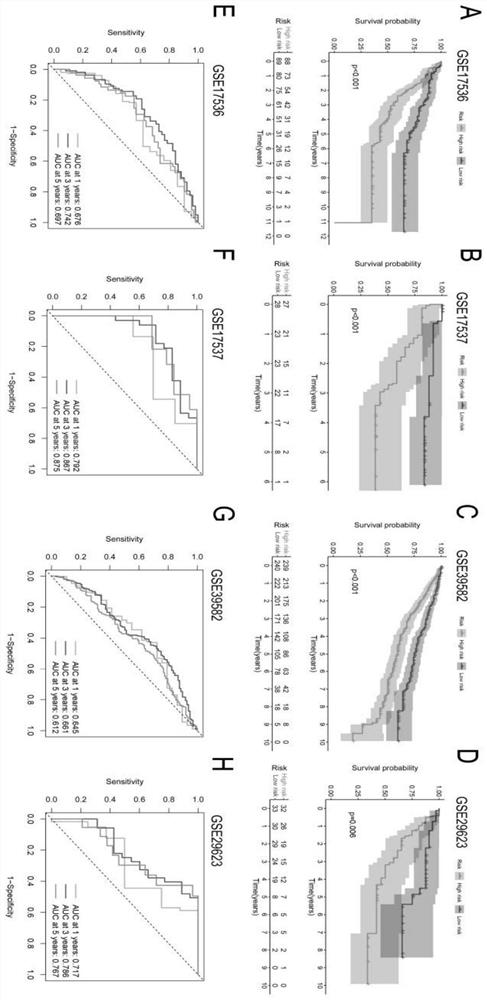
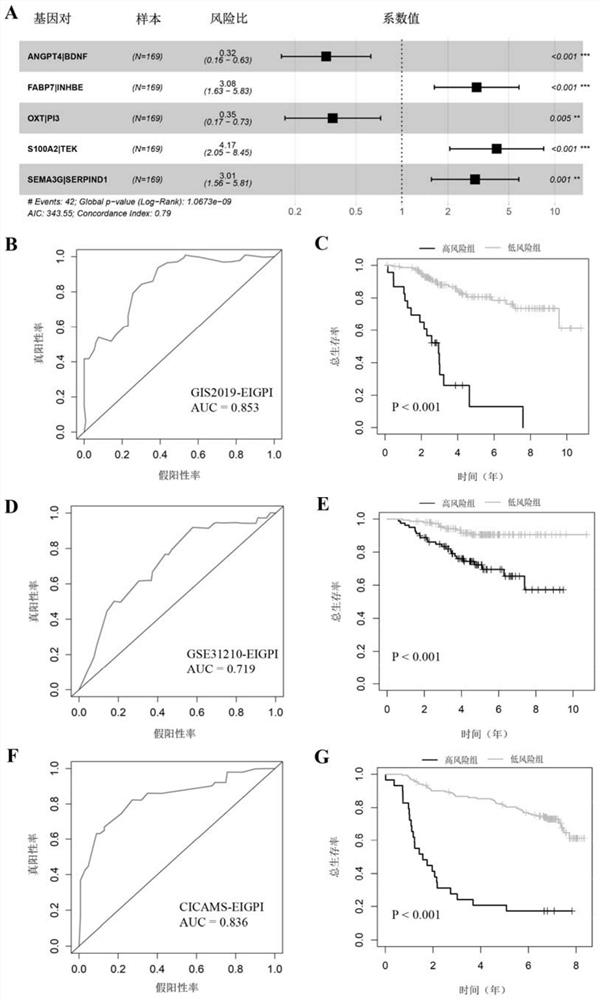
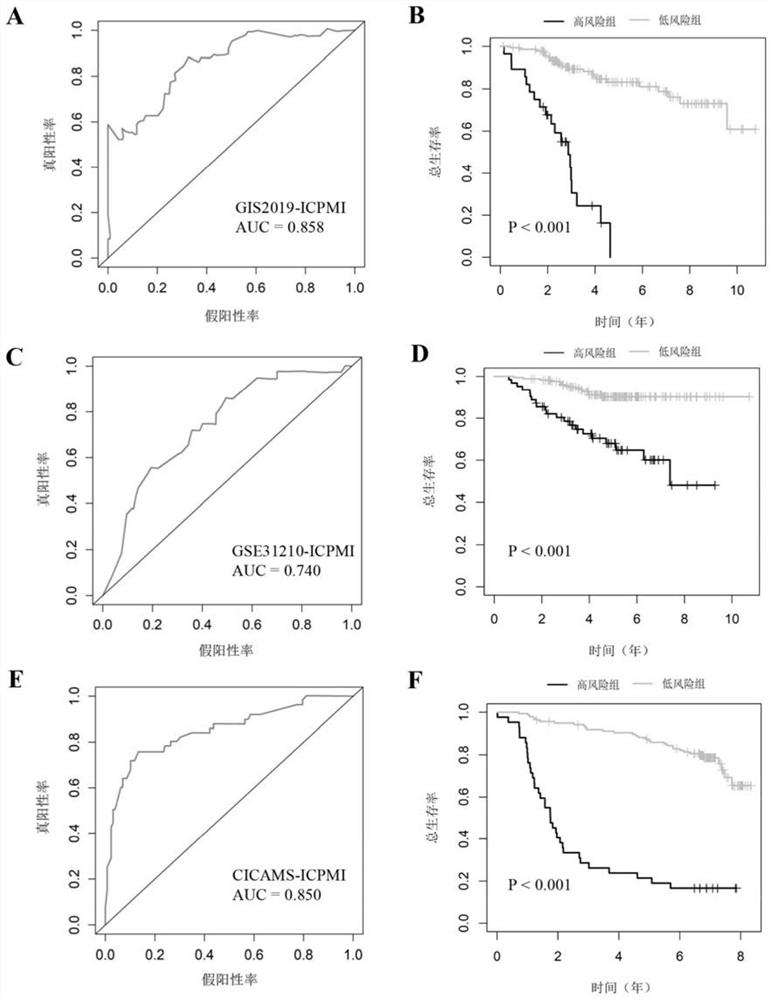
The invention discloses an immune-clinical feature combined prediction model for evaluating prognosis of lung adenocarcinoma. The invention provides a prognosis risk prediction system for Asian lung adenocarcinoma patients. The prognosis risk prediction system comprises a system for detecting expression quantities of ten genes, namely ANGPT4, BDNF, FABP7, INHBE, OXT, PI3, S100A2, TEK, SEMA3G and SERPIND1, of tumor samples of the Asian lung adenocarcinoma patients. According to the invention, the immune-clinical prognosis model index is systematically researched for the first time. According to the research, the relationship among EGFR mutation, immunophenotype and prognosis of Asian lung adenocarcinoma patients is systematically discussed for the first time, and a composite immune and clinical model related to EGFR mutation is established. The model may be a reliable and promising prognosis tool, and is helpful for further personalized management of patients.
Owner:CANCER INST & HOSPITAL CHINESE ACADEMY OF MEDICAL SCI
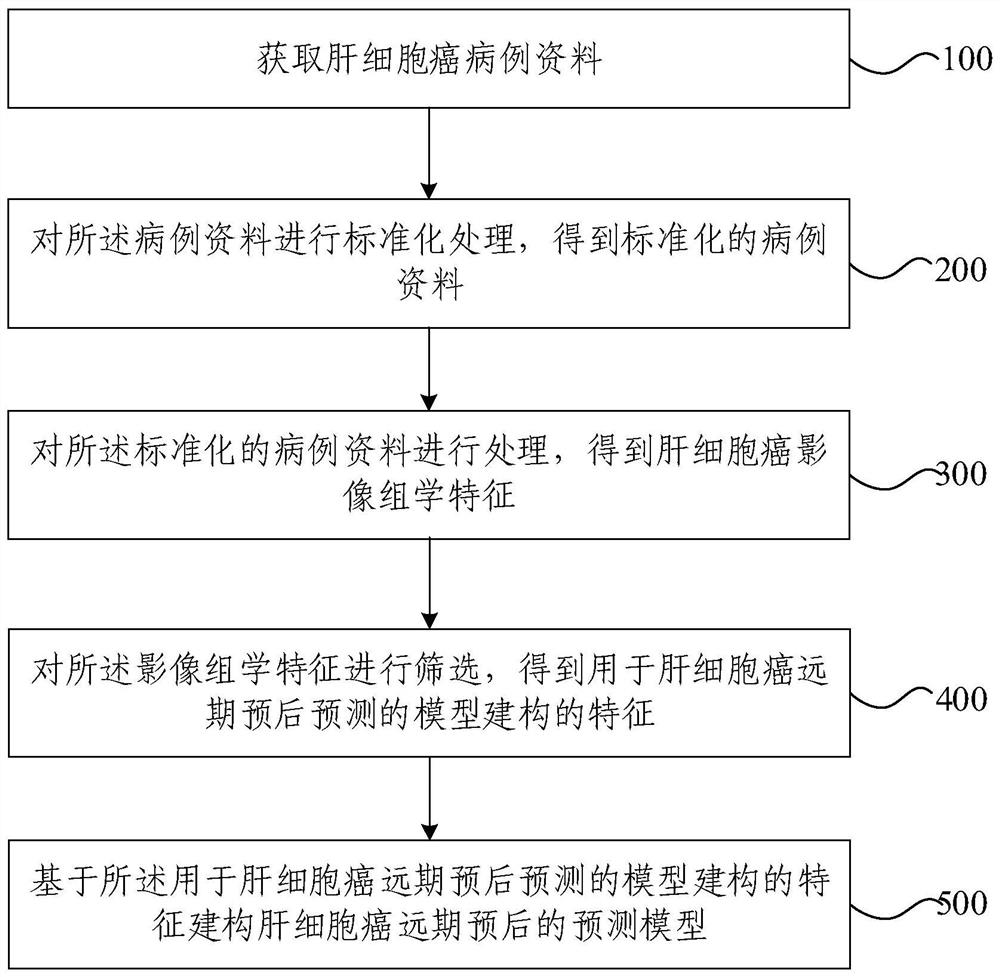
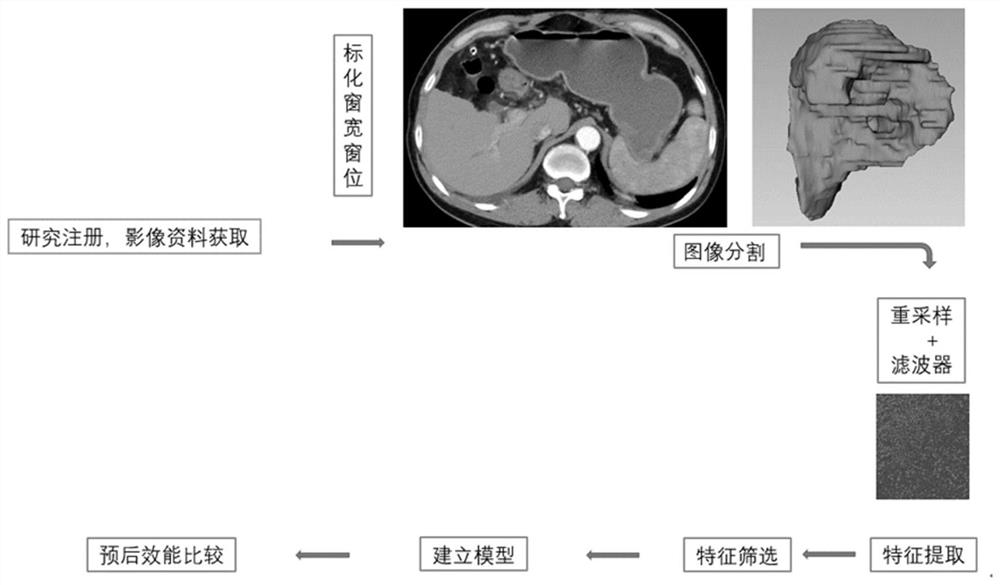
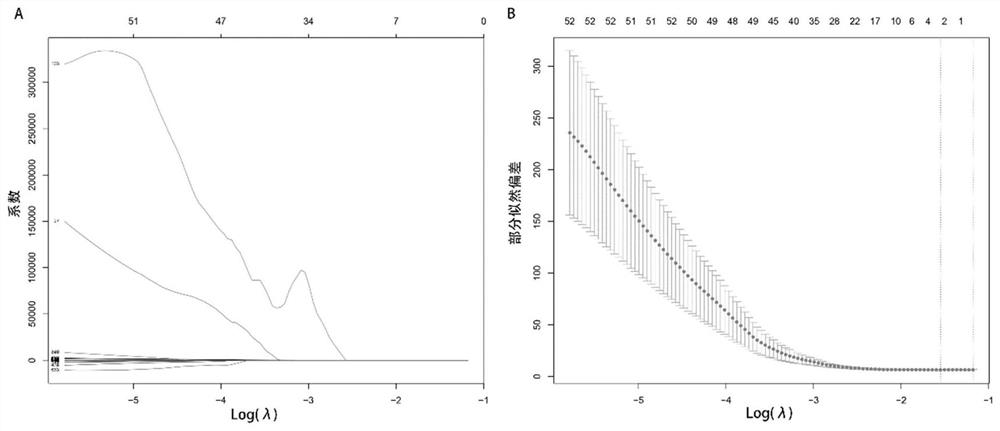
Modeling method and device for long-term prognosis prediction of hepatocellular carcinoma
ActiveCN113571194AEarly therapeutic interventionMedical simulationMedical data miningHepatocellular carcinomaPrognosis prediction
The invention provides a modeling method and device for long-term prognosis prediction of hepatocellular carcinoma. The method comprises the steps of obtaining hepatocellular carcinoma case data; performing standardization processing on the case data to obtain standardized case data; processing the standardized case data to obtain hepatocellular carcinoma radiomics characteristics; screening the radiomics characteristics to obtain characteristics for constructing a model for long-term prognosis prediction of hepatocellular carcinoma; and constructing a prediction model for long-term prognosis of hepatocellular carcinoma on the basis of the characteristics for construction of the model for long-term prognosis prediction of hepatocellular carcinoma. The postoperative image characteristics are screened according to the long-term prognosis of hepatocellular carcinoma, and the prognosis model is established according to the screened characteristics, so that accurate prediction of the long-term prognosis of hepatocellular carcinoma and advanced treatment intervention are realized, and support of whole-process image data is provided for the long-term prognosis prediction of hepatocellular carcinoma.
Owner:TSINGHUA UNIV
Prognostic risk assessment system for gastric cancer patients
PendingCN112908477APredict Prognostic SurvivalExcellent sensitivityHealth-index calculationProteomicsGastric carcinomaDifferential Methylation
A prognosis risk assessment system for gastric cancer patients is characterized by comprising a gastric cancer methylation data acquisition module, a differential methylation site acquisition module and a prognosis model construction module. The gastric cancer methylation data acquisition module is used for acquiring TCGA gastric cancer methylation spectrum and a gastric cancer methylation spectrum data set GSE30601, the differential methylation site acquisition module is used for obtaining a plurality of hyper-methylation sites and a plurality of hypo-methylation sites, and the prognosis model construction module is used for constructing a 11-DNA methylation site risk scoring formula of each patient.
Owner:黑龙江省医院
Method for constructing diagnostic model of prostatic cancer
The invention discloses a method for constructing a diagnostic model of prostatic cancer. The method comprises the following steps: Step 1) obtaining gene expression profile data of PCa; 2, differential expression profile analysis is conducted on the gene expression profile data of the PCa, and differential genes in the PCa are screened out; 3, key genes are screened through a GAE algorithm in a machine learning method for differential genes in PCa; step 4) carrying out PPI analysis on a calculation result of the GAE to obtain 10 high-expression genes and 6 low-expression genes; 5, a prognosis model is established through single-factor regression analysis and multi-factor regression analysis; 6, a diagnosis model of the PCa is constructed according to the prognosis model parameters; and (7) the PCa diagnosis model is verified. According to the invention, the PCa diagnosis model constructed by four genes is constructed and verified, so that a basis is provided for personalized precise treatment of PCa patients.
Owner:CHONGQING MEDICAL UNIVERSITY
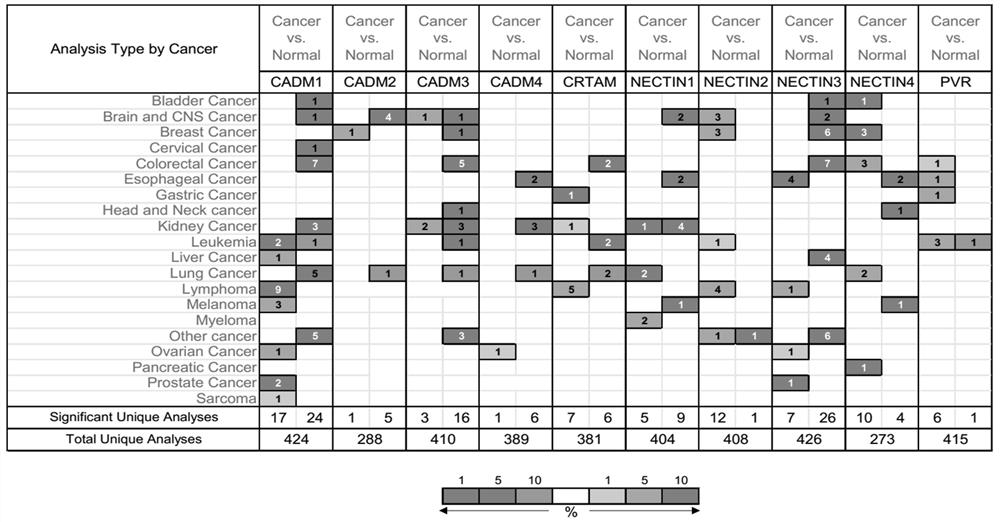
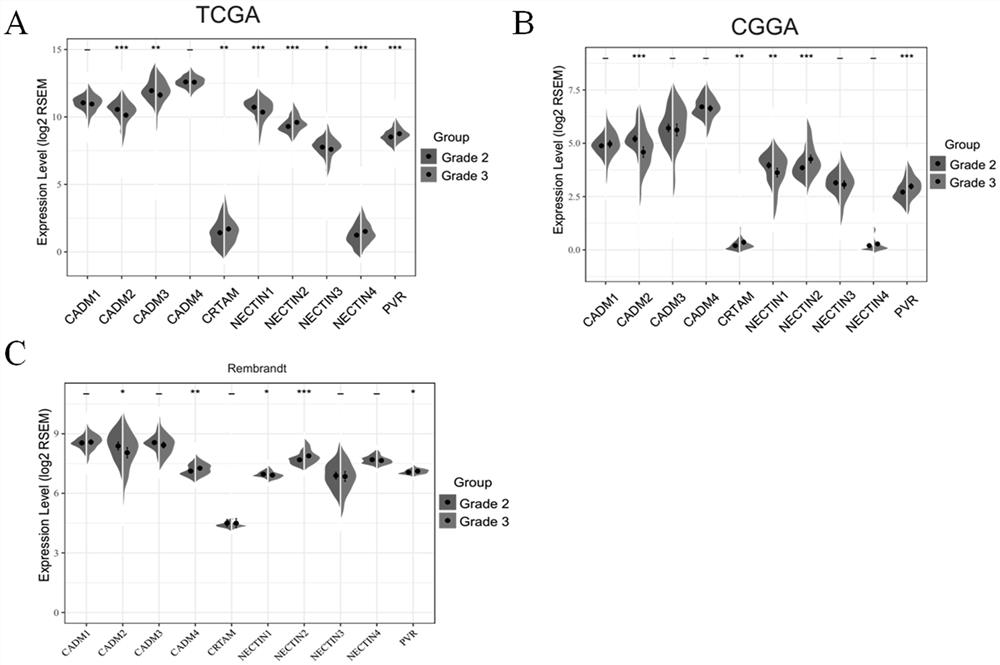
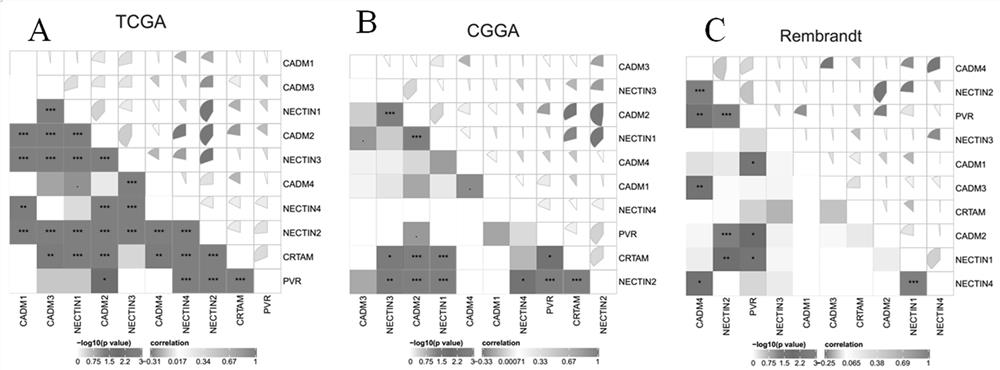
Application of NECTIN family and NECTIN-like molecules in low-level glioma prognosis model
The invention belongs to the technical field of biological information and biomedicine, and particularly relates to application of NECTIN family and NECTIN-like molecules in a low-grade glioma prognosis model. By evaluating expression profiles in the three independent databases, four prognosis related genes CADM2, NECTIN1, NECTIN2 and PVR between the nectin and the necls are determined, and the genes have huge potential as LGG treatment targets and prognosis biomarkers. In addition, the invention also discusses a potential pathway participating in new four-gene risk scoring, and finds that the risk scoring model plays an important role in the immune process of LGG. The invention provides wide nectin and Necls analysis, provides a four-gene model for prognosis prediction of the LGG, provides a thought for further researching CADM2, NECTIN1 / 2 and PVR as potential clinical and immune targets of the LGG, and has wide clinical application panorama.
Owner:THE FIRST HOSPITAL OF CHINA MEDICIAL UNIV
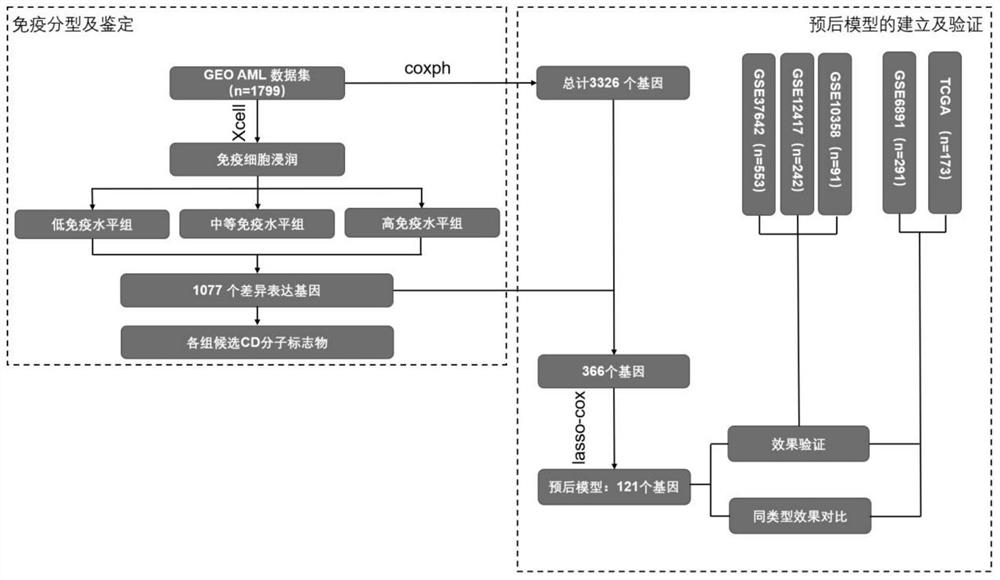
AML patient immune typing system, AML patient prognostic scoring model and construction method thereof
ActiveCN113053456AImprove forecast accuracySignificant difference in prognosisHealth-index calculationEpidemiological alert systemsClinical prognosisCell type
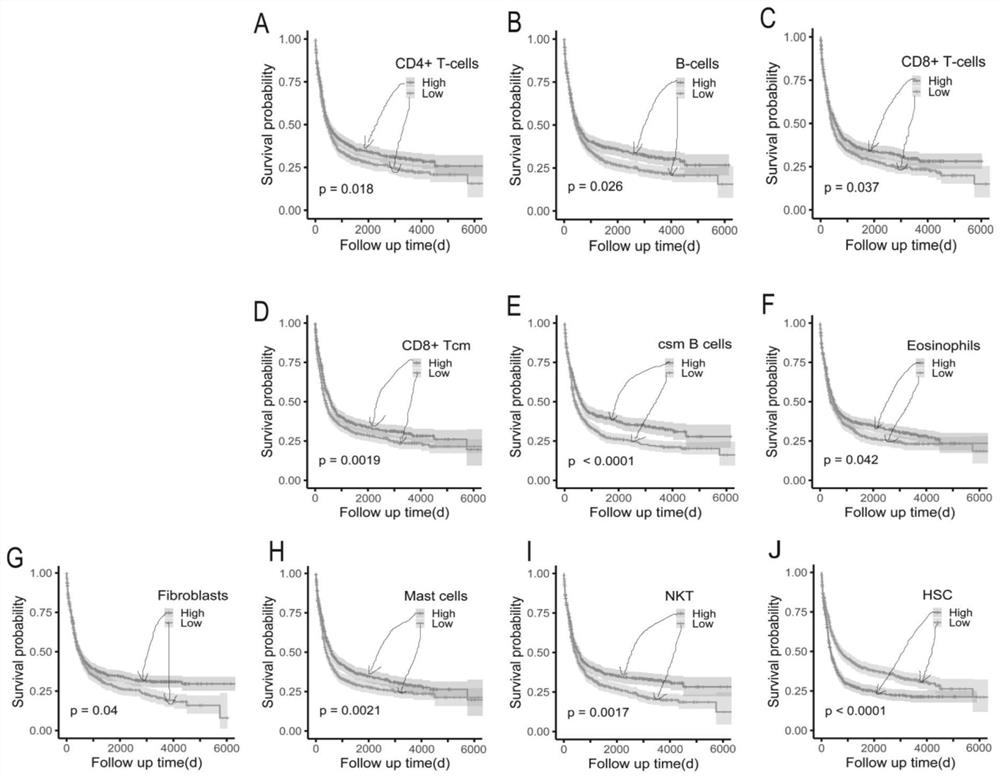
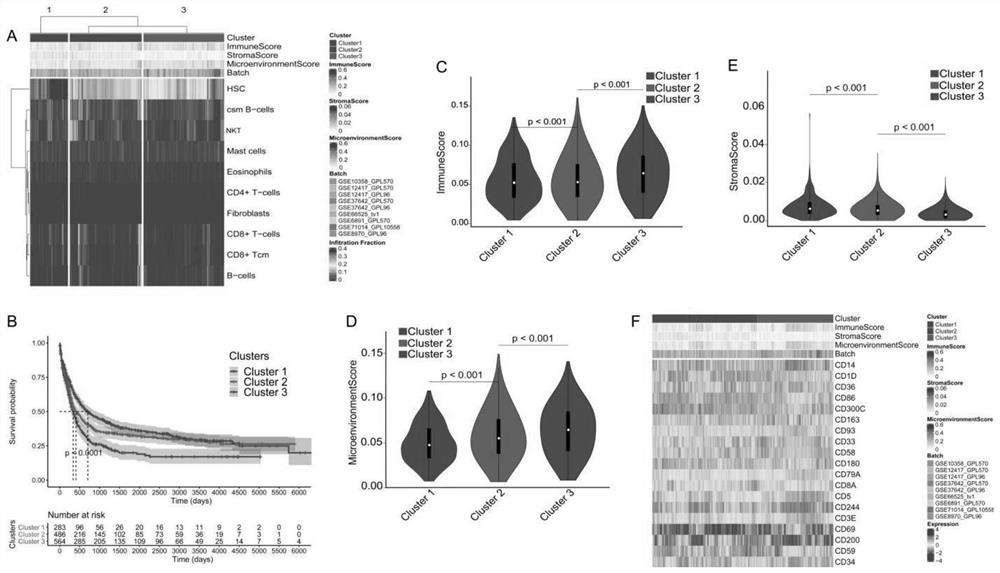
The invention discloses an AML patient immune typing system, an AML patient prognosis scoring model and a construction method thereof. The construction method of the immune typing system comprises the following steps: 1) collecting immune microenvironment information of an AML patient from a GEO database; 2) carrying out clustering analysis to obtain 10 immune cell types with clinical prognosis significance; 3) clustering the patients into three groups: Cluster1, Cluster2 and Cluster3, the immune score and the microenvironment score of the Cluster1 being the lowest, and the HSC infiltration degree of the Cluster1 being the highest; the immune score and the microenvironment score of the Cluster3 being the highest, and the HSC infiltration degree of the Cluster3 being the lowest; the Cluster2 being between the two groups, so that the establishment of the immune typing system is completed. In addition, an AML patient prognosis model is constructed according to the immune typing system, and the model is higher in precision compared with an existing model.
Owner:THE SECOND AFFILIATED HOSPITAL OF GUANGZHOU MEDICAL UNIV
Brain tumor radiotherapy mode intelligent selection method, system, equipment and medium
PendingCN114171157AImprove experienceImprove the efficiency of diagnosis and treatmentMechanical/radiation/invasive therapiesCharacter and pattern recognitionFeature vectorImaging analysis
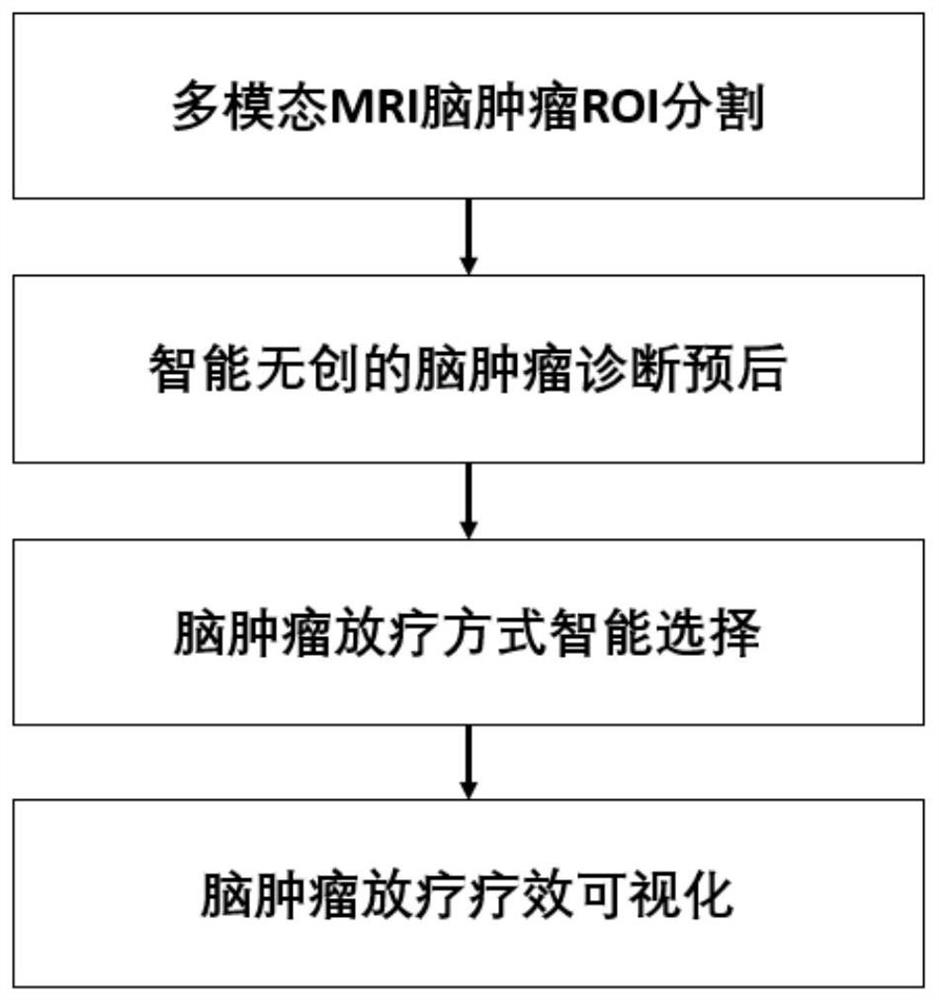
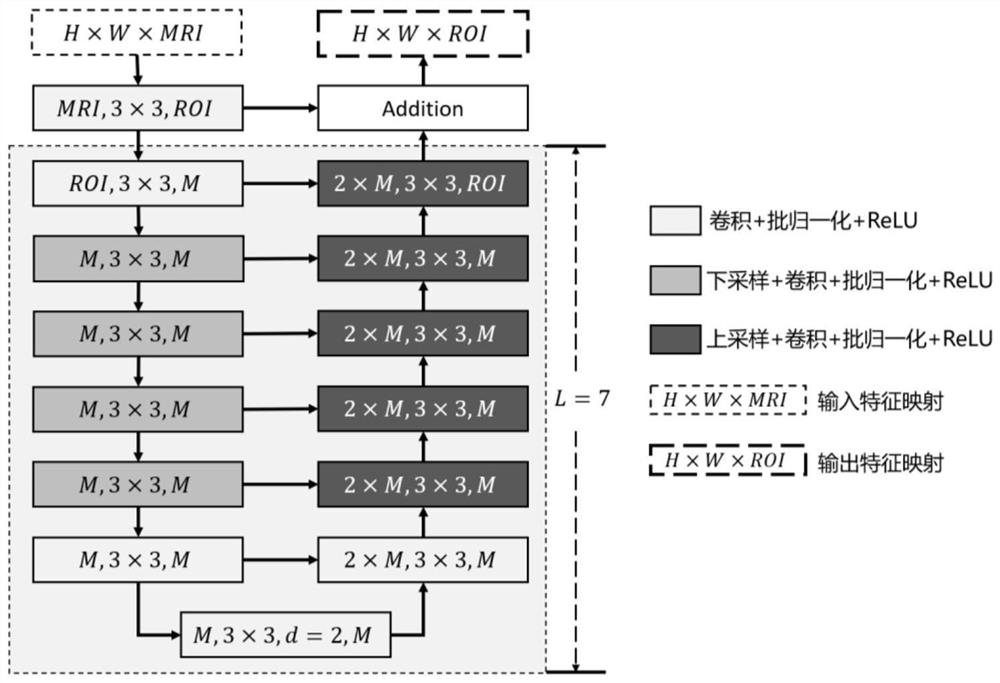
The invention relates to a brain tumor radiotherapy mode intelligent selection method, system and device and a medium, and the method comprises the steps: constructing a multi-mode MRI brain tumor segmentation model, carrying out the brain tumor ROI segmentation of a multi-mode MRI image before and after radiotherapy of a patient, and obtaining a brain tumor pre-radiotherapy region and a brain tumor post-radiotherapy residual region which are paired; constructing a brain tumor diagnosis prognosis model, and taking the effective feature vectors as input to obtain prediction results of pathological grading and radiotherapy prognosis; constructing a brain tumor radiotherapy mode intelligent selection model, and taking image gene features and radiotherapy sensitive features before radiotherapy as input to obtain an optimal radiotherapy mode; and constructing a brain tumor radiotherapy curative effect visual model, and obtaining a predicted post-radiotherapy MRI image by taking the brain tumor MRI image of the patient before radiotherapy, the prediction results of pathological grading and radiotherapy prognosis and the optimal radiotherapy mode as input. The method can be widely applied to the medical image analysis technology and the application field.
Owner:INST OF MODERN PHYSICS CHINESE ACADEMY OF SCI
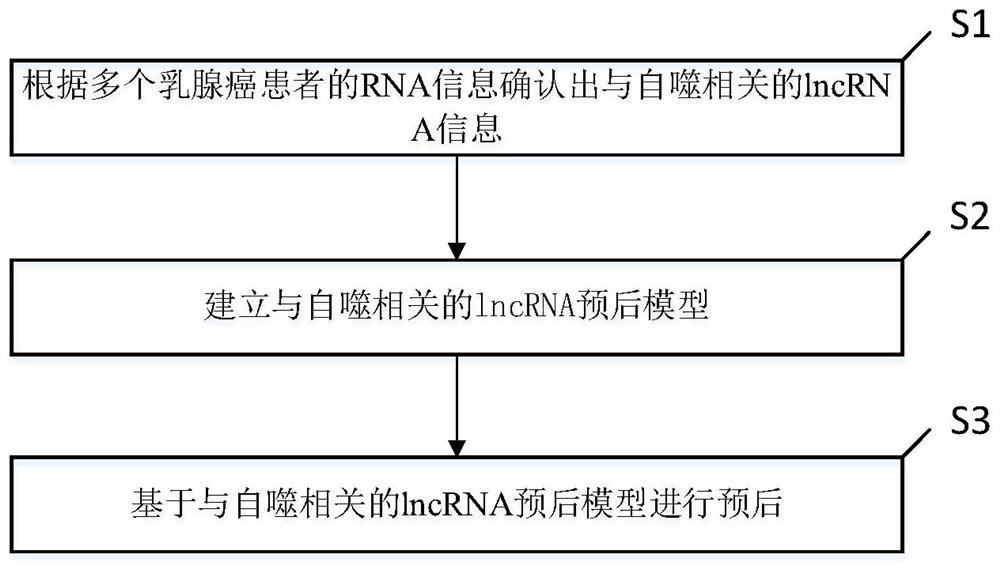
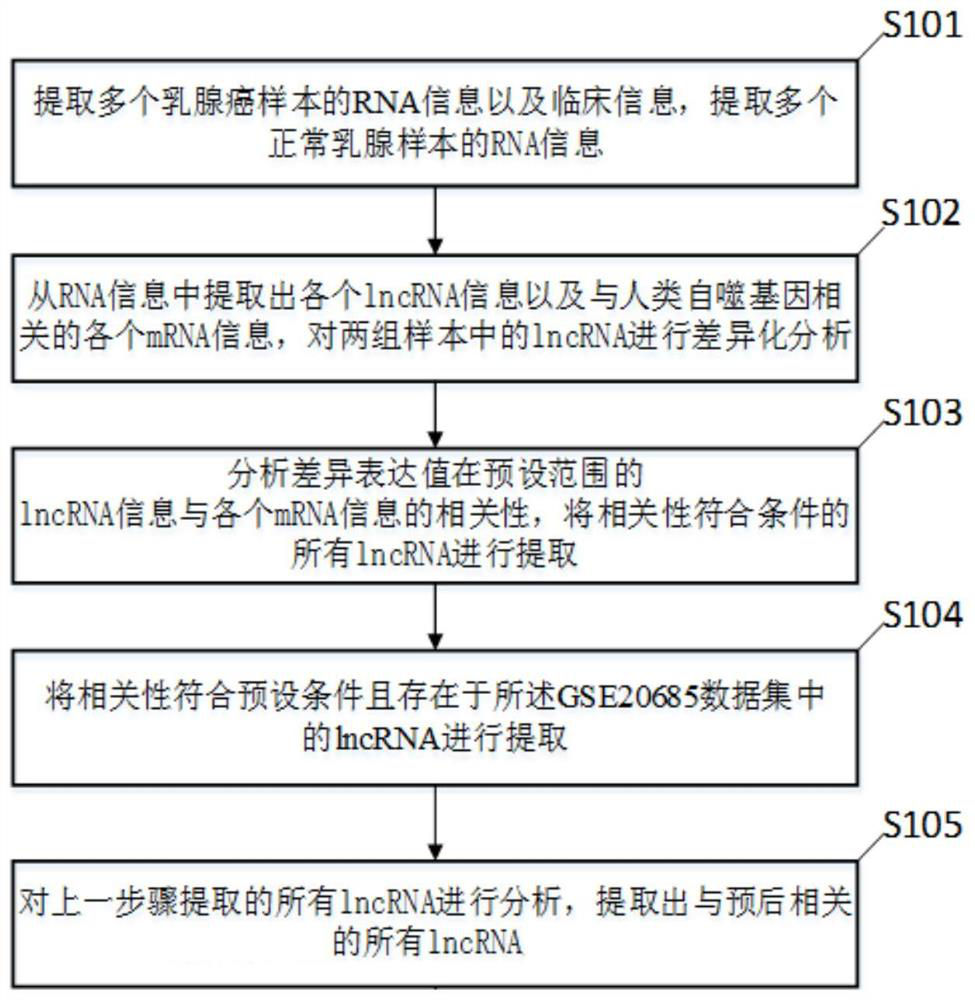
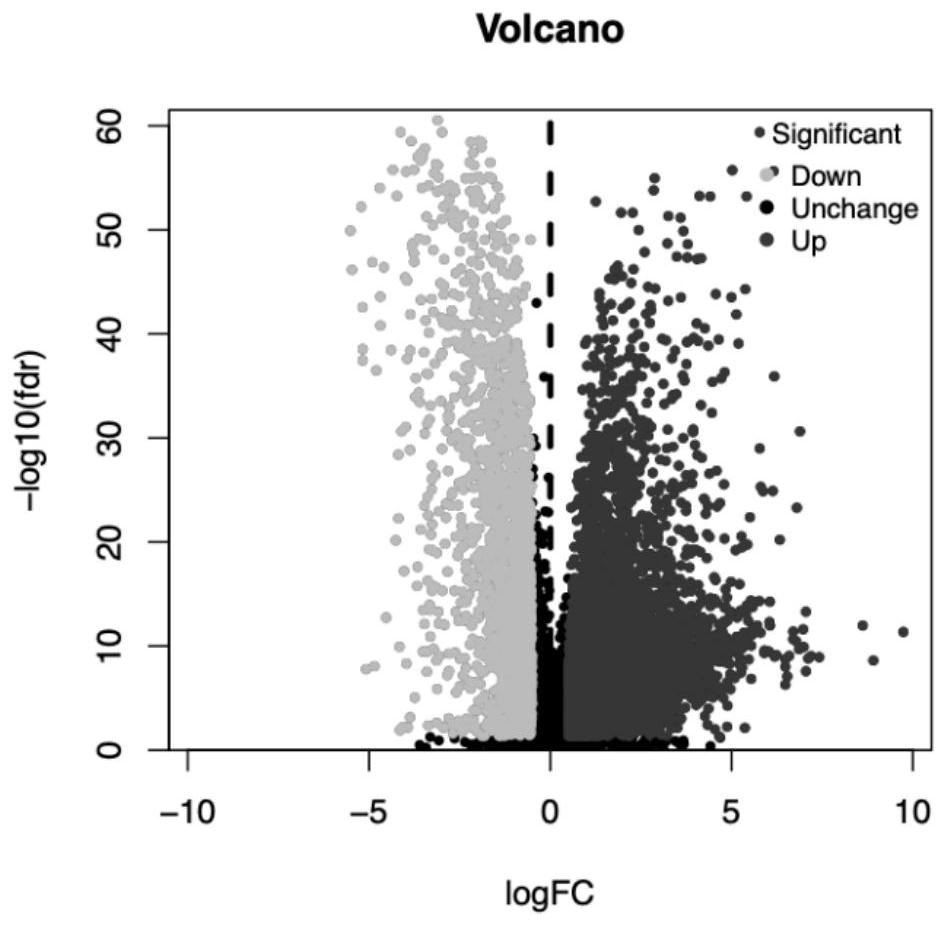
Breast cancer prognosis evaluation method and system based on autophagy related lncRNA model
PendingCN113593648AUniversalImprove prognostic accuracyHealth-index calculationBiostatisticsCancer researchHuman Mammary Glands
The invention discloses a breast cancer prognosis evaluation method and system based on an autophagy related lncRNA model, and the breast cancer prognosis evaluation method based on the autophagy related lncRNA model is used for prognosis of breast cancer. The breast cancer prognosis evaluation method based on the autophagy-related lncRNA model comprises the following steps: determining autophagy-related lncRNA information according to RNA information of a plurality of breast cancer samples and RNA information of a plurality of normal breast samples; analyzing the lncRNA information related to the autophagy and the clinical data of the plurality of breast cancer samples, and establishing an lncRNA prognosis model related to the autophagy; and performing prognosis based on the autophagy-related lncRNA prognosis model. According to the breast cancer prognosis evaluation method and system based on the autophagy related lncRNA model, the prognosis of the breast cancer sample can be accurately judged.
Owner:ZHENGZHOU UNIV
Breast cancer molecular typing and risk assessment method based on mRNA expression
PendingCN113652485AReduce the impact of accuracyImprove accuracyMicrobiological testing/measurementProteomicsLuminal bRNA extraction
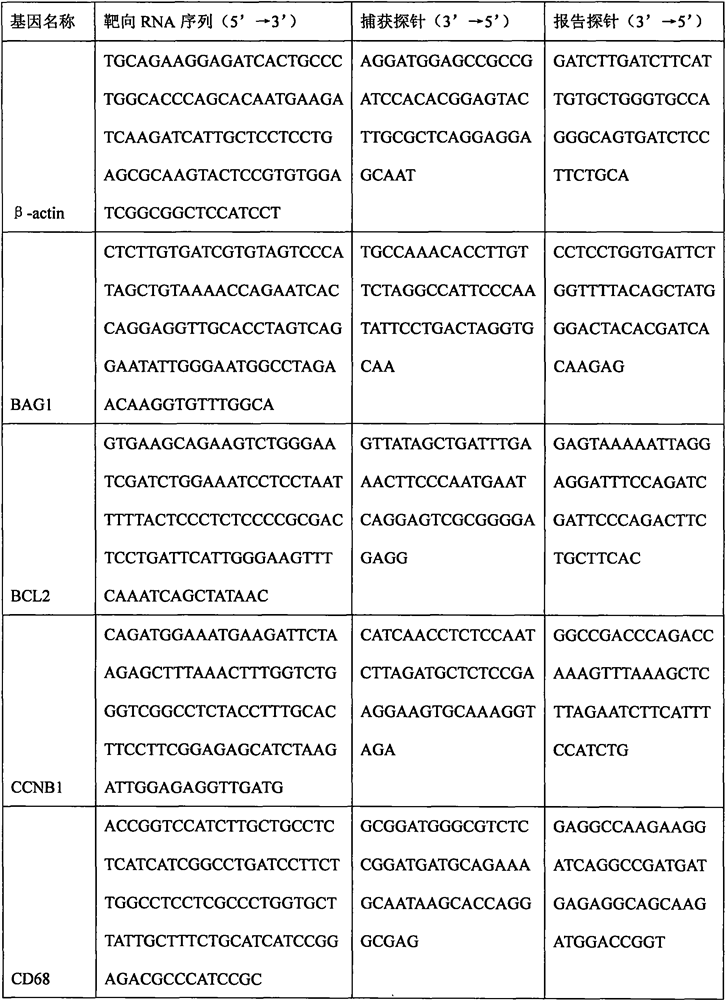
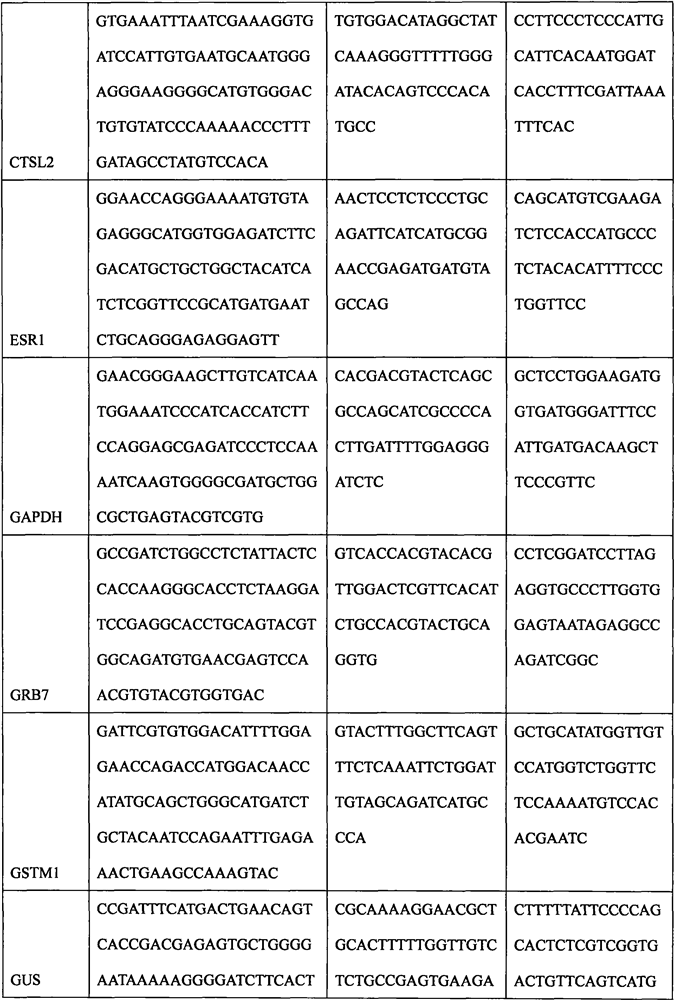
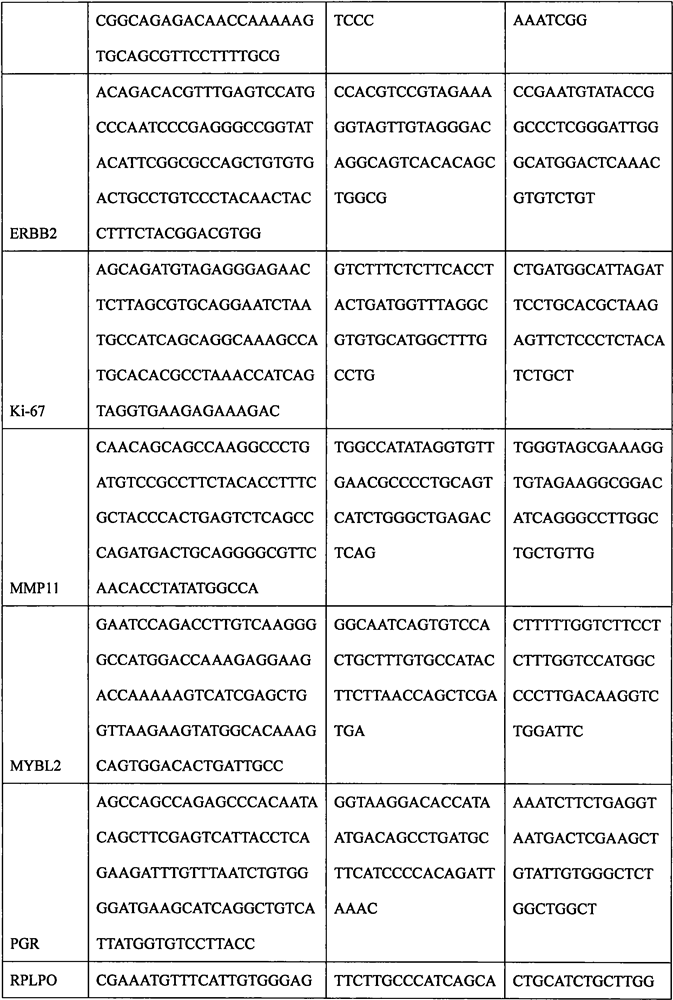
The invention discloses a breast cancer molecular typing and risk assessment method based on mRNA expression, and particularly relates to the technical field of medical treatment. The method comprises the following specific steps: step 1, extracting total RNA in a breast cancer paraffin specimen by adopting a Qiagen FFPE RNA extraction kit; 2, measuring the total RNA concentration. The method comprises the following steps: carrying out hybridization reaction on an mRNA quantitative probe and a target RNA sequence, sealing a film on a hybridization reaction product, putting the hybridization reaction product into a nanoString digital fluorescent bar code single-molecule analyzer, and carrying out gene expression detection through a program set in advance; then carrying out quantitative analysis on the expression quantities of the HER2 gene, the ESR1 gene, the PGR gene and the Ki-67 gene by adopting nSolver software; then, various specimens are subjected to luminal type molecular typing, HER2 + type molecular typing and triple-negative type molecular typing according to set thresholds, luminal types are further divided into luminal A type molecular typing and luminal B type molecular typing and HER2 type molecular typing are further divided into luminal B-HER2 + type molecular typing and HER2 + type molecular typing according to the threshold of CTSL2, and an ER + positive prognosis model is established, so that low-risk people can be accurately predicted, and treatment is avoided.
Owner:HENAN CANCER HOSPITAL
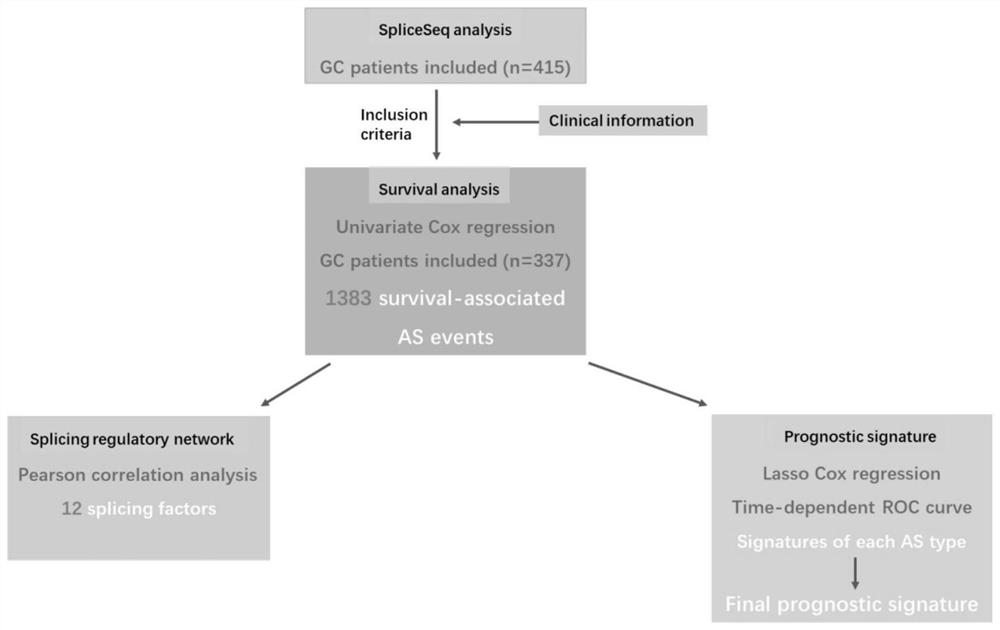
Construction method and application of gastric cancer prognosis model based on alternative splicing events
PendingCN111899889APredicted survivalMedical data miningHealth-index calculationGastric carcinomaAlternative splicing
The invention belongs to the technical field of biomedicine and medical informatics, and particularly relates to a construction method and application of a gastric cancer prognosis model based on alternative splicing events. According to the invention, a sample is reasonably selected, alternative splicing events significantly related to the total lifetime of patients with gastric cancer are determined through single-factor Cox regression analysis, and then the prognosis model is established by using a minimum absolute constriction and selection operator (LASSO) and multi-factor Cox analysis; and the final prognosis model based on seven types of alternative splicing events can be used as an independent prognosis index of a patient with gastric cancer.
Owner:GUIZHOU MEDICAL UNIV
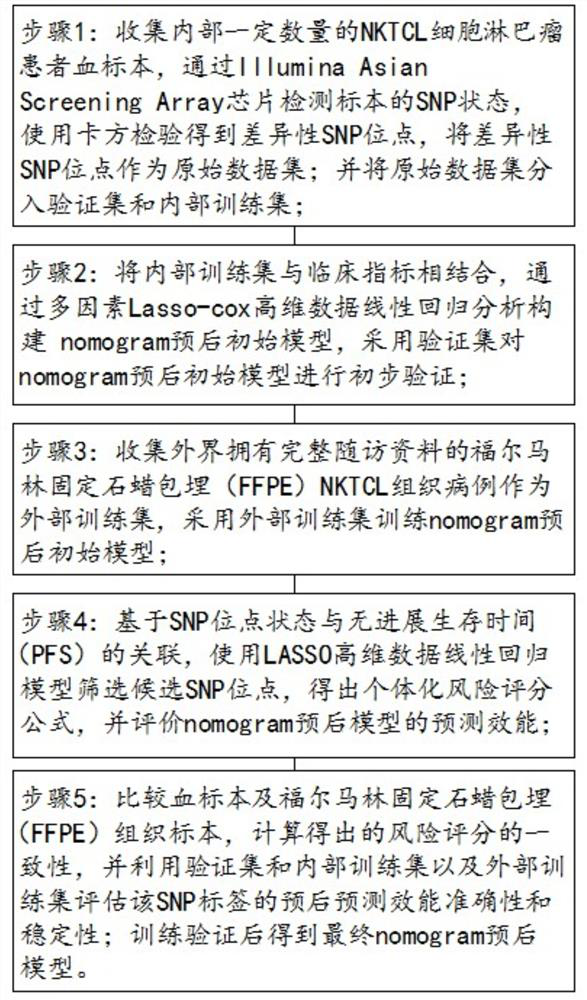
Construction method of NKT cell lymphoma individualized prognosis model based on mononucleotide
PendingCN114645093ALow cost of treatmentReduce risk of recurrenceMechanical/radiation/invasive therapiesHealth-index calculationPrognostic modelsLinear regression
The invention specifically discloses a construction method of an NKT cell lymphoma individualized prognosis model based on mononucleotide. The construction method comprises the following steps: collecting a certain number of internal blood specimens of NKTCL cell lymphoma patients, detecting the SNP state of the specimens through an Illumina Asian Screening Array chip, obtaining differential SNP sites by using chi-square test, and taking the differential SNP sites as an original data set; dividing the original data set into a verification set and an internal training set; the internal training set is combined with clinical indexes, a nomogram prognosis initial model is constructed through multi-factor Lasso-cox high-dimensional data linear regression analysis, and the nomogram prognosis initial model is preliminarily verified through a verification set; the method comprises the following steps: collecting an external formalin-fixed paraffin embedding (FFPE) NKTCL tissue case with complete follow-up visit data as an external training set, and training a nomogram prognosis initial model by adopting the external training set; and the initial model is predicted, and a final nomogram prognosis model is obtained after verification. The method can guide precise treatment to save medical resources, relieve the burden of a patient and reduce the treatment side reaction of the patient.
Owner:SUN YAT SEN UNIV CANCER CENT
Marker genes for oocyte competence
ActiveUS20180327841A1Health-index calculationMicrobiological testing/measurementMammalPotential biomarkers
Cumulus cell (CC) gene expression is being explored as an additional method to morphological scoring to choose the embryo with the highest chance to pregnancy. The present invention relates to a novel method of identifying biomarker genes for evaluating the competence of a mammalian oocyte in giving rise to a viable pregnancy after fertilization, based on the use of live birth and embryonic development as endpoint criteria for the oocytes to be used in an exon level analysis of potential biomarker genes. The invention further provides CC-expressed biomarker genes thus identified, as well as prognostic models based on the biomarker genes identified using the methods of the present invention.
Owner:VRIJE UNIV BRUSSEL
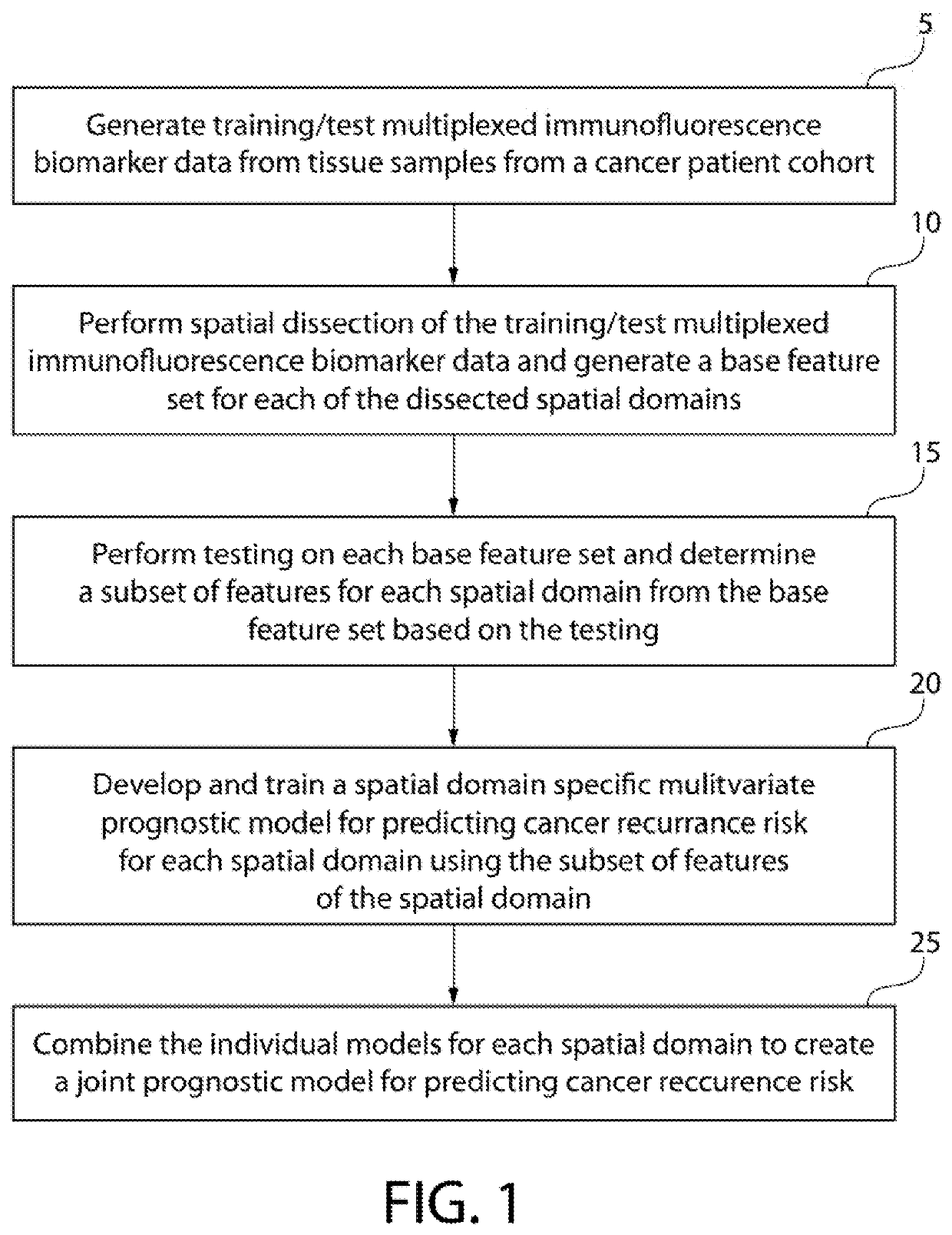
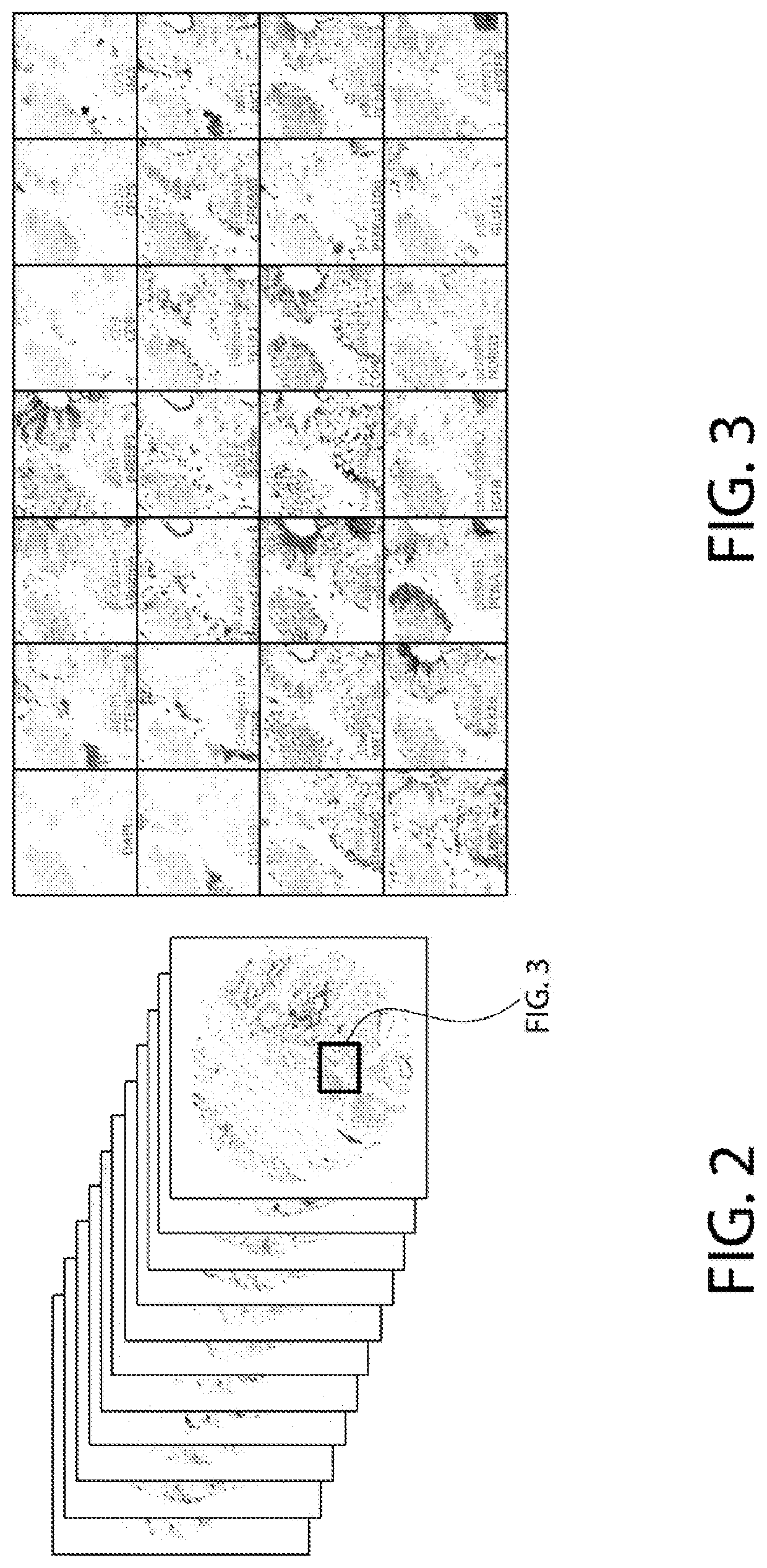
Predicting cancer recurrence from spatial multi-parameter cellular and subcellular imaging data
A method of predicting cancer recurrence risk for an individual includes receiving patient spatial multi-parameter cellular and sub-cellular imaging data for a tumor of the individual, and analyzing the patient spatial multi-parameter cellular and sub-cellular imaging data using a prognostic model for predicting cancer recurrence risk to determine a predicted cancer recurrence risk for the individual, wherein the joint prognostic model is based on spatial correlation statistics among features derived for a plurality of intra-tumor spatial domains from spatial multi-parameter cellular and sub-cellular imaging data obtained from a plurality of cancer patients.
Owner:UNIVERSITY OF PITTSBURGH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com