Application of model constructed based on PCD related gene combination in preparation of product for predicting prognosis of colonic adenocarcinoma
A colon adenocarcinoma and gene combination technology, applied in the field of biomedicine, can solve the problem of unclear effect of cancer immunotherapy, achieve optimal immune response and tolerance, avoid overtreatment and delay the patient's condition.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
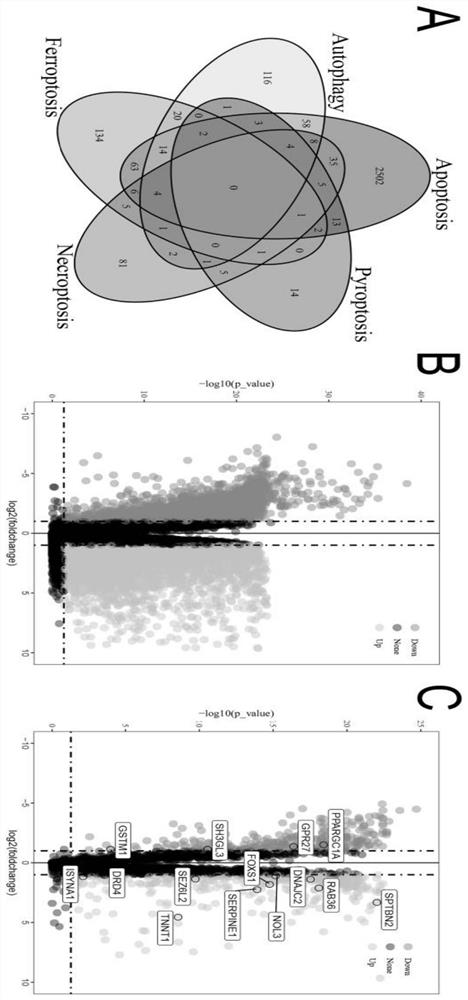
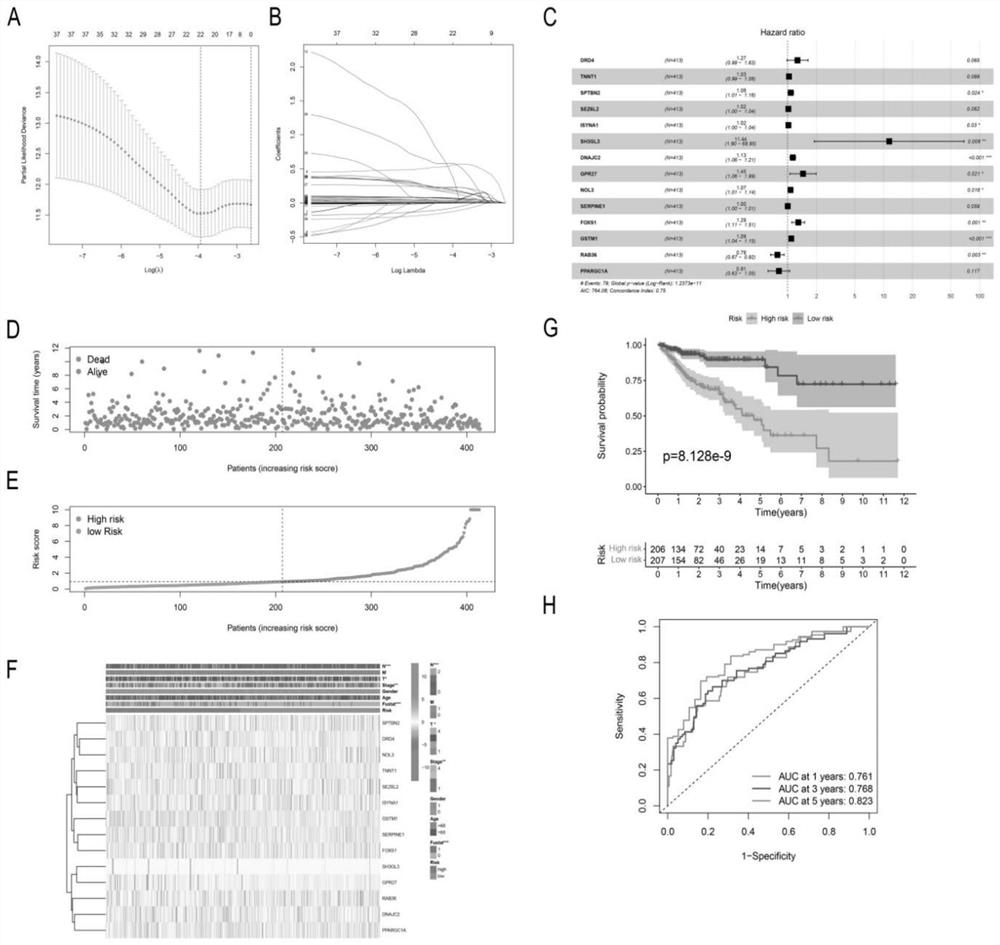
[0028] Screening of PCD-related genes for the construction of a model for COAD prognostic analysis.
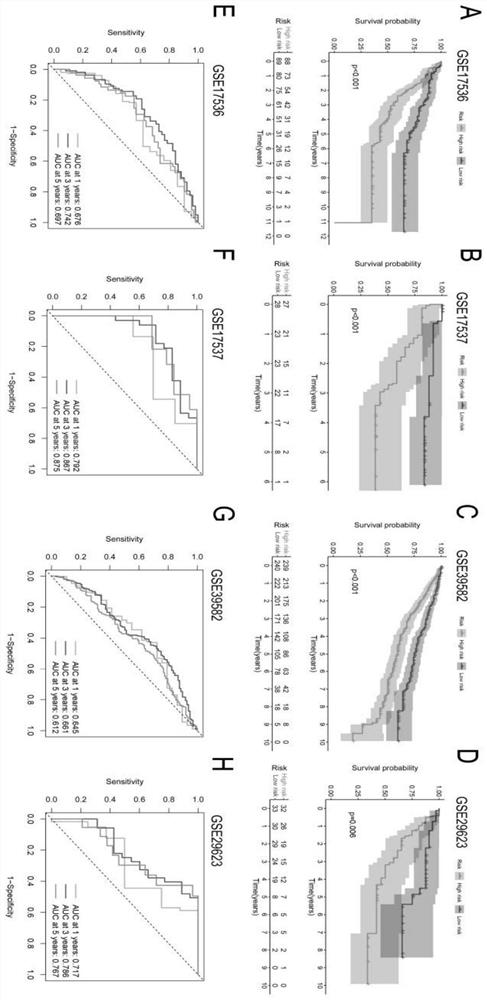
[0029] (1) Obtaining patient information: RNA-seq data and clinical characteristics of COAD patients were obtained from The Cancer Genome Atlas (TCGA) database (https: / / portal.gdc.cancer.gov / reposito57Sry). After data screening and filtering, COAD samples and normal samples were selected from the TCGA-COAD cohort. Four external validation datasets GSE17536 (n=177), GSE17537 (n=55), GSE39582 (n=479) and GSE29623 (n=65) were used for external validation. As shown in Table S1.
[0030] (2) Screening prognosis-related PCD-related genes as prognostic biomarkers for constructing a prognostic model and validating the prognostic model: use
[0031] http: / / www.zhounan.org / ferrdb / , http: / / www.autophagy.lu / index.html http: / / www.gsea-msigdb.org / gsea / index.jsp, https: / / www.kegg .jp / entry / hsa04217 and other databases screened and extracted ferroptosis, autophagy, pyroptosis, apoptosis an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com