Second-generation sequencing library building method for identifying bacterial types and drug-resistant types of helicobacter pylori in samples
A technology of Helicobacter pylori and second-generation sequencing, which is applied in the fields of biochemical equipment and methods, chemical libraries, and microbial determination/inspection, can solve the problems of cumbersome operation procedures, expensive reagents, and low upper limit of the number of target genes. high cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0056] Example: Identification of drug-resistant types and bacterial typing of Helicobacter pylori in stool samples
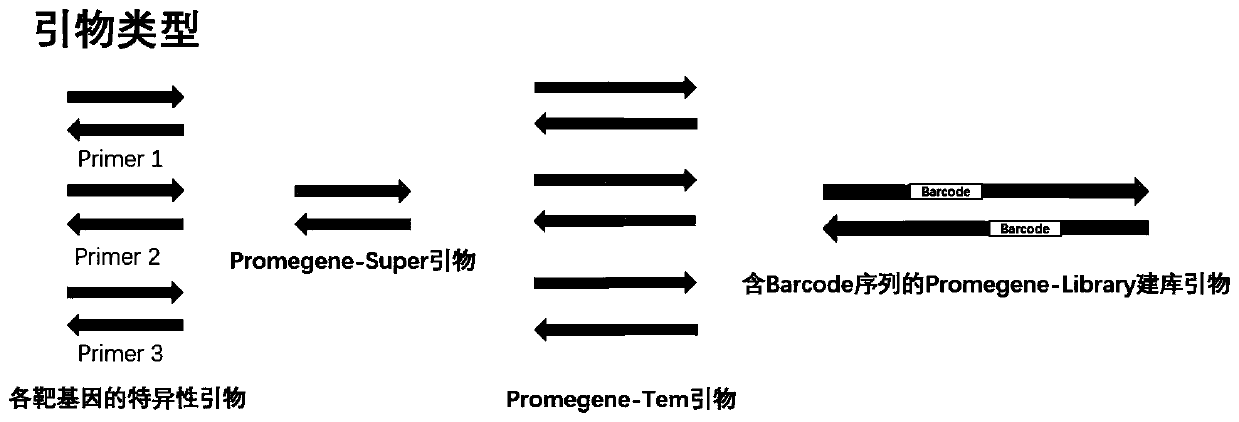
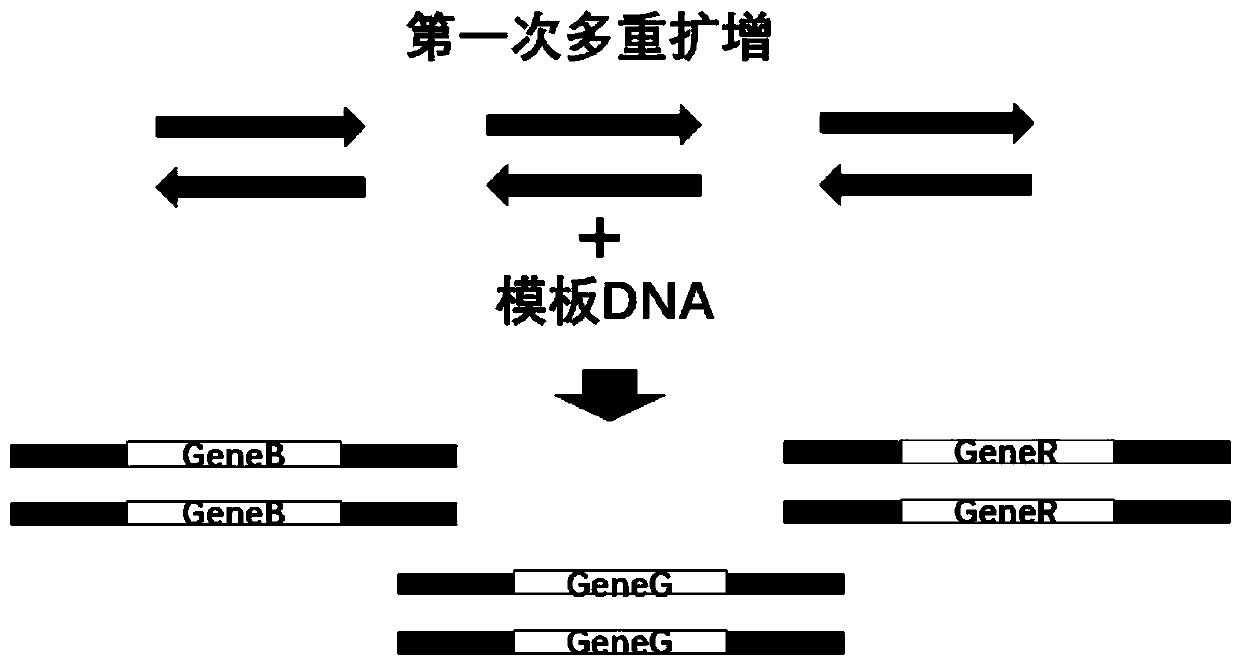
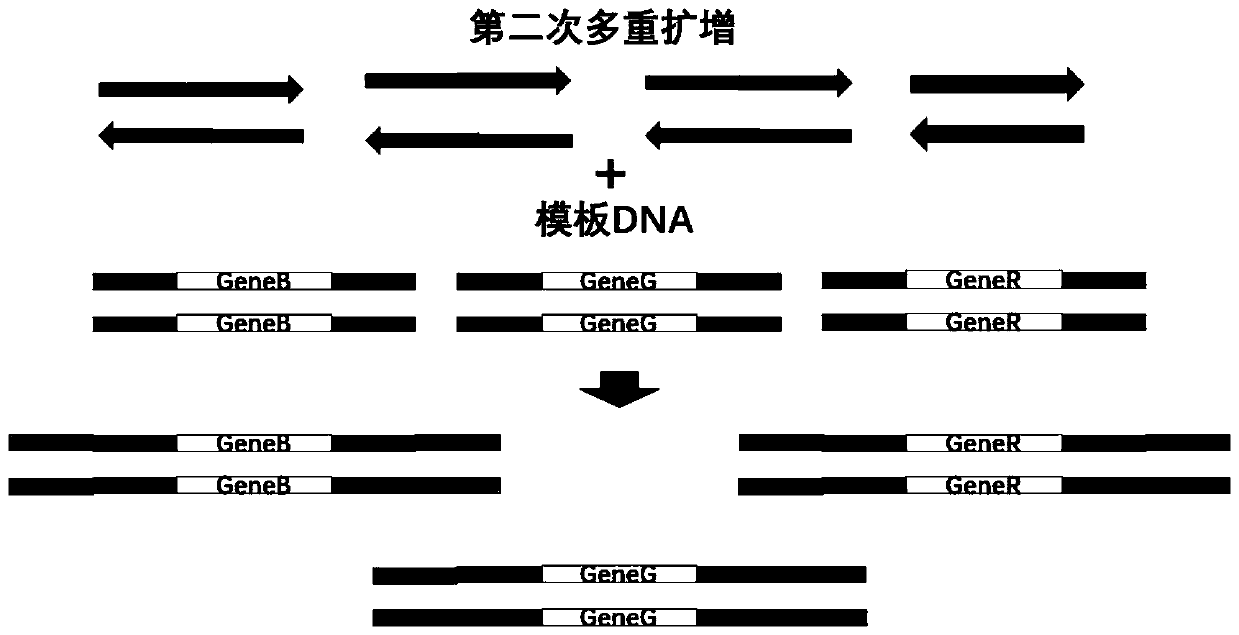
[0057] By reviewing relevant kit products, patents and literature, it was determined that 3 related genes involved in 2 types of H. pylori - UreA gene, VacA gene and CagA gene, and 9 thermal mutation sites related to five drug resistance Points and 5 related genes of 15 mutation types—RdxA gene, 23S gene, Pbp1 gene, GyrA gene and 16S gene.
[0058] After determining the target gene to be detected, the location of the drug-resistant mutation site of each target gene was considered, and primers and alternative primers were designed for each target gene, so that the sequencing length of the three genes for typing was 300bp Within (to ensure that the full length of the gene can be measured), the mutation sites of 5 drug resistance-related genes are located within 150 bp from the start of the two ends of the sequencing (to ensure that at least one end of each gene c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com