Zearalenone hydrolase mutant ZHDM1, and encoding gene and application thereof
A technology of zearalenone and ZHDM1, applied in the field of agricultural biology, can solve the problems of low activity and hinder large-scale industrial production and application of zearalenone hydrolase, and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Preparation of recombinant strain X33 (pPICZ(α)A-zh607) containing wild-type erythralenone hydrolase gene
[0032] 1.1 Amplify the nucleic acid sequence zh607 of the wild-type ZH607 of the zearalenone hydrolase protein
[0033] After codon optimization, the coding gene zh607 of erythralenone hydrolase ZH607 was obtained. The PCR method was used to amplify the zh607 gene and the vector pPICZ(α)A nucleic acid fragment, and connect the two through a recombination kit to obtain the recombinant plasmid pPICZ(α)A-zh607, and transform Pichia pastoris X33 to obtain recombinant Pichia pastoris Strain X33 (pPICZ(α)A-zh607).
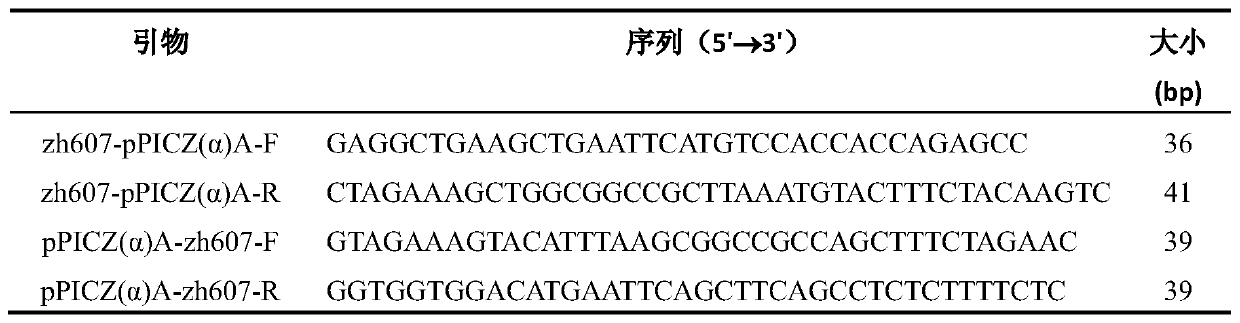
[0034] The primers used in PCR are as follows:
[0035] Table 1 Wild-type zearalenone hydrolase and specific primers for vector amplification
[0036]
[0037] Among them, zh607-pPICZ(α)A-F and zh607-pPICZ(α)A-R are used to amplify the gene coding sequence of wild type ZH607 of zearalenone hydrolase; pPICZ(α)A-zh607-F and pPICZ(α)A -zh607-R ...
Embodiment 2
[0042] Example 2 Preparation of recombinant strain X33 (pPICZ(α)A-zh607-AH) containing mutant zearalenone hydrolase gene
[0043] 2.1 Construction of recombinant plasmid pPICZ(α)A-zh607-AH
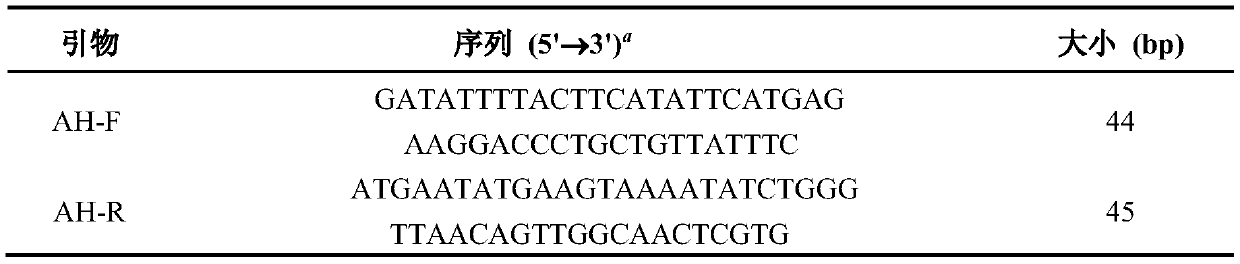
[0044] The optimized mutation site was designed to replace ASVTGME at position 136-142 with DILLHIH, and the mutation site was introduced by the method of point mutation kit, and it was sequenced and verified to obtain the chiaralenone hydrolase mutant plasmid pPICZ(α) A-zh607-AH. The primers used are shown in Table 2:
[0045] Table 2 Specific primers for mutant zearalenone hydrolase
[0046]
[0047] 2.2 Construction of recombinant strain X33 (pPICZ(α)A-zh607-AH)
[0048] The recombinant plasmid pPICZ(α)A-zh607-AH was digested with SacI, and the recovered product was transformed into Pichia pastoris competent cells X33 by electroporation to induce expression, and the recombinant expression strain X33 (pPICZ(α)A-zh607-AH) was obtained.
Embodiment 3
[0049] Example 3 Obtaining wild-type ZH607 and mutant ZHDM1 of erythralenone hydrolase protein
[0050] 3.1 Induced expression of protein ZH607 and ZHDM1
[0051] The obtained recombinant expression strains X33 (pPICZ(α)A-zh607) and X33(pPICZ(α)A-zh607-AH) were inoculated into YPD medium for seed culture, 200rpm, 30°C for 48h, and 1% The inoculum was transferred to BMGY medium, cultured at 200 rpm at 30°C for 48 hours, and after enough bacteria were enriched, the bacteria were collected and added to BMMY medium containing 1% methanol to induce expression.
[0052] 2. Purification of protein ZH607 and ZHDM1
[0053] Centrifuge the induced bacterial solution at 12,000 rpm for 10 minutes, collect the supernatant and concentrate it, then dialyze it with 10 mM disodium hydrogen phosphate solution (adjust the pH to 7.6 with citric acid), and then carry out ion exchange chromatography on the dialyzed enzyme solution. Solution A is 10mM disodium hydrogen phosphate solution (adjust t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Specific vitality | aaaaa | aaaaa |
| Specific vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com