Method, kit and oligonucleotide for detecting novel coronavirus
An oligonucleotide, coronavirus technology, applied in the field of life science and biology, to achieve fast and objective test results, high sensitivity, and avoid false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Extraction of clinical sample nucleic acid
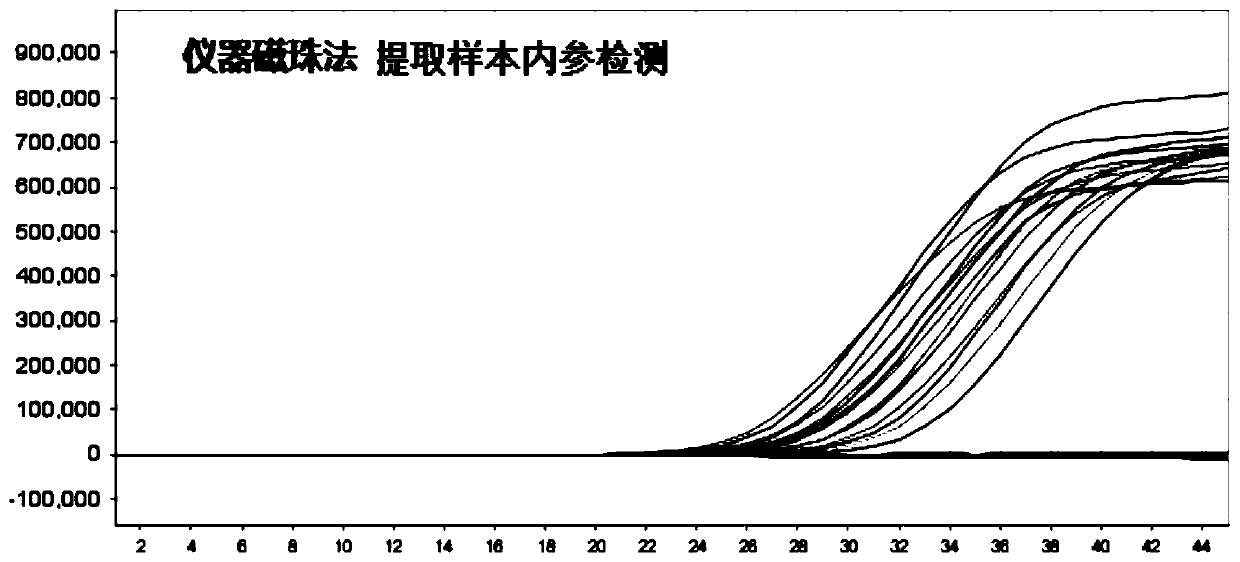
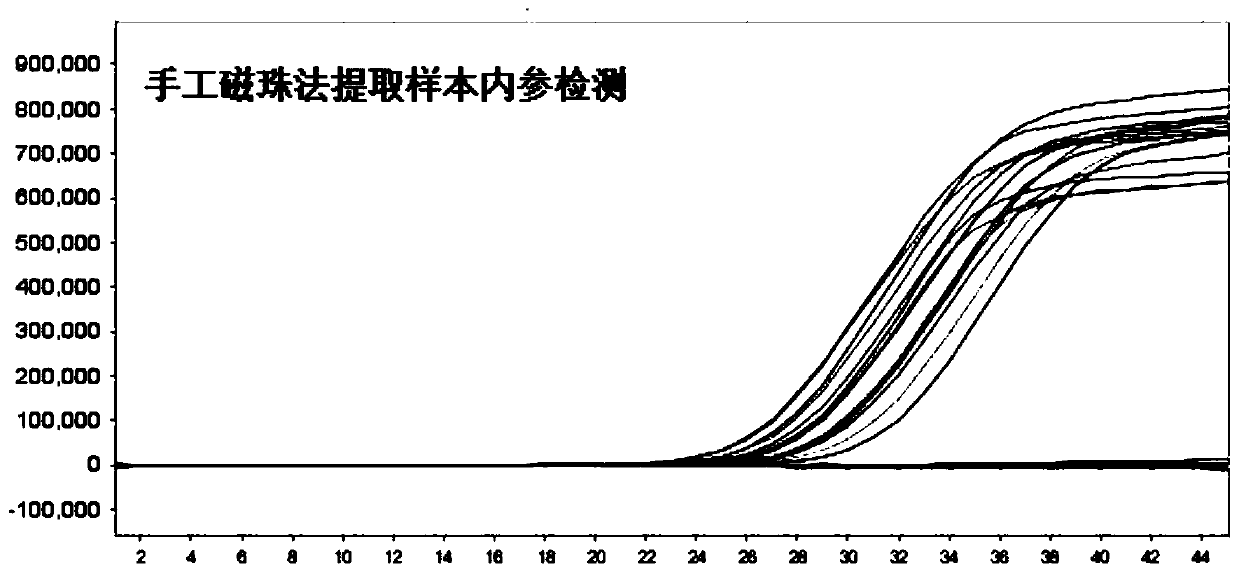
[0065] For the convenience of illustration, in this example, normal oral and pharyngeal swabs were used as clinical samples, and the manual magnetic bead method described in the present invention was used to extract nucleic acid from clinical samples, and the extraction kit of the instrumental magnetic bead method was used as a control for comparison.
[0066]Get 10 cases of normal oral and pharyngeal swabs, and extract sample RNA according to the three-step method of the present invention: the first step is to get 0.5mL sample and 0.5mL nucleic acid extraction solution 1, let it stand still for 10min, discard the supernatant after the magnetic frame absorbs the magnetic beads; Add 400 μL of nucleic acid extraction solution 2 in the first step, vortex and mix well, and then stand still for 5 minutes. The magnetic stand absorbs the magnetic beads and discards the supernatant; the third step adds 100 μL of nucleic ...
Embodiment 2
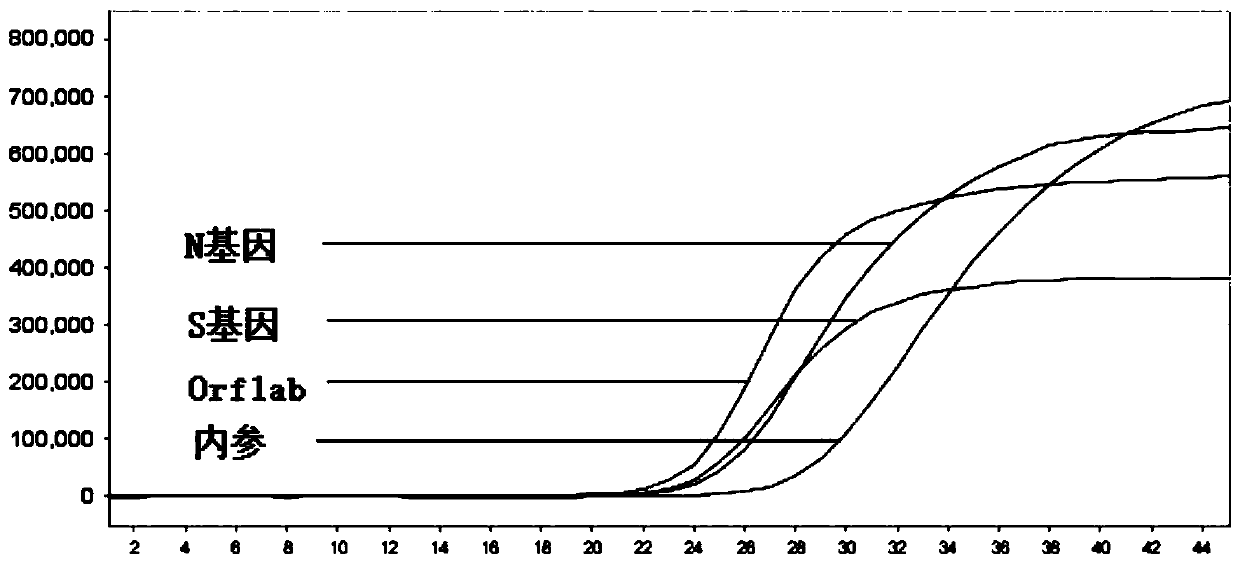
[0070] Example 2: Detection of novel coronavirus by fluorescent RT-PCR method
[0071] In this example, a kit is used for fluorescent RT-PCR detection of novel coronavirus, and the kit includes fluorescent RT-PCR reaction solution, enzyme mixture, positive quality control, and negative quality control. Preferably, this kit can also include the nucleic acid extraction solution 1, the nucleic acid extraction solution 2 and the nucleic acid extraction solution 3 in Example 1. The preparation system of fluorescent RT-PCR reaction solution is shown in Table 3. The enzyme mixture preparation system is shown in Table 4. The positive quality control is the new coronavirus SARS-CoV-2 pseudovirus, and the negative quality control is 0.9% NaCl solution.
[0072] Table 3. Fluorescence RT-PCR reaction liquid preparation system table (1 person)
[0073]
[0074]
[0075] Table 4. Enzyme mixture preparation system (1 serving)
[0076]
[0077]
[0078] When detecting the path...
Embodiment 3
[0095] Embodiment 3: clinical sample verification
[0096] In the clinical center, 290 cases of nucleic acid extraction samples were tested for clinical evaluation.
[0097] According to the method described in Example 2, the novel coronavirus fluorescent RT-PCR reaction solution preparation system was prepared, and the 290 nucleic acid extraction samples were subjected to fluorescent RT-PCR amplification, and the test results were recorded. Among these 290 samples, 118 cases were positive and 172 cases were negative (as control results) as determined by the clinical center combining molecular detection, CT imaging and epidemiological investigation results. The analysis of the detection results of the present invention for the three target genes is shown in Table 8.
[0098] Table 8. Clinical verification results of novel coronavirus nucleic acid detection
[0099]
[0100] Positive coincidence rate = [115 / (3+115)] × 100% = 97.46%
[0101] Negative coincidence rate = [16...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com