Method for identifying disease-related miRNA and method for deducing enrichment signal pathway
A technology for signaling pathways and diseases, applied in the field of identification of disease-causing genes, which can solve the problems of incompleteness, inability of prediction tools to discover miRNAs, and difficulty in information.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0049] The present invention will be further elaborated below according to the drawings and specific embodiments.
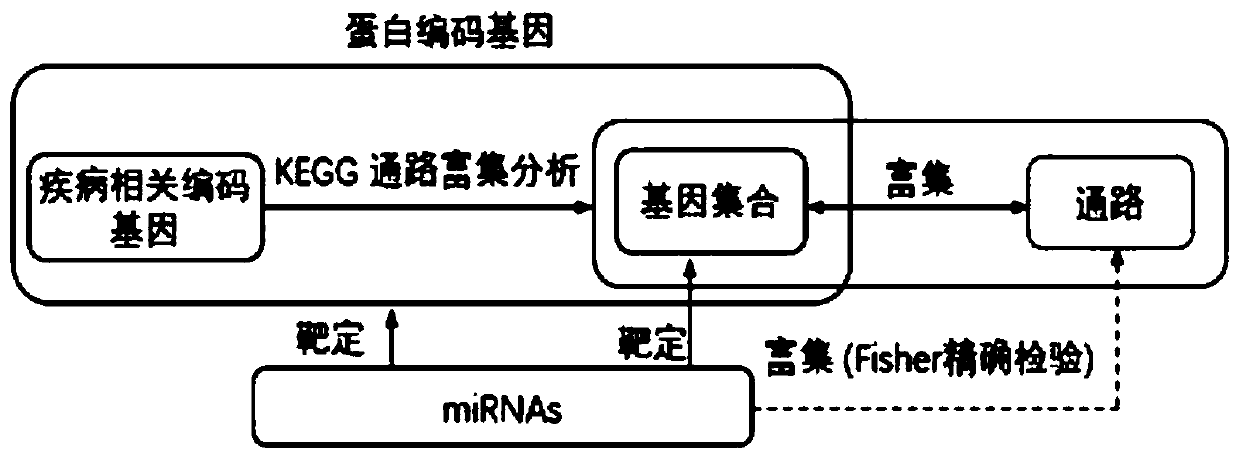
[0050] The present invention is a method for inferring the enrichment signal pathway of disease-associated miRNA, specifically according to the following steps:
[0051] Step 1. Use the PRISMA process to collect data set A of a specific disease-related miRNA:
[0052] Such as figure 1 As shown, all the literatures of disease-related miRNAs were obtained from the three databases of Medline, PubMed, and Embase. After removing duplicate literatures, in the remaining literatures, review the titles and abstracts of the articles one by one to exclude inappropriate literatures, and then review The full text of the literature excludes inappropriate literature to obtain the required miRNA data set;
[0053] Inappropriate literature mentioned above includes literature not related to humans or mice, not related to a specific disease, case reports, and non-original researc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com