A method for determining homologous recombination defects based on DNA sequencing data
A DNA sequencing and homologous recombination technology, applied in the field of data science, can solve problems such as large errors, undetermined HRD status, difficult determination and optimization of judgment thresholds, etc., to achieve good judgment results and overcome data insufficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
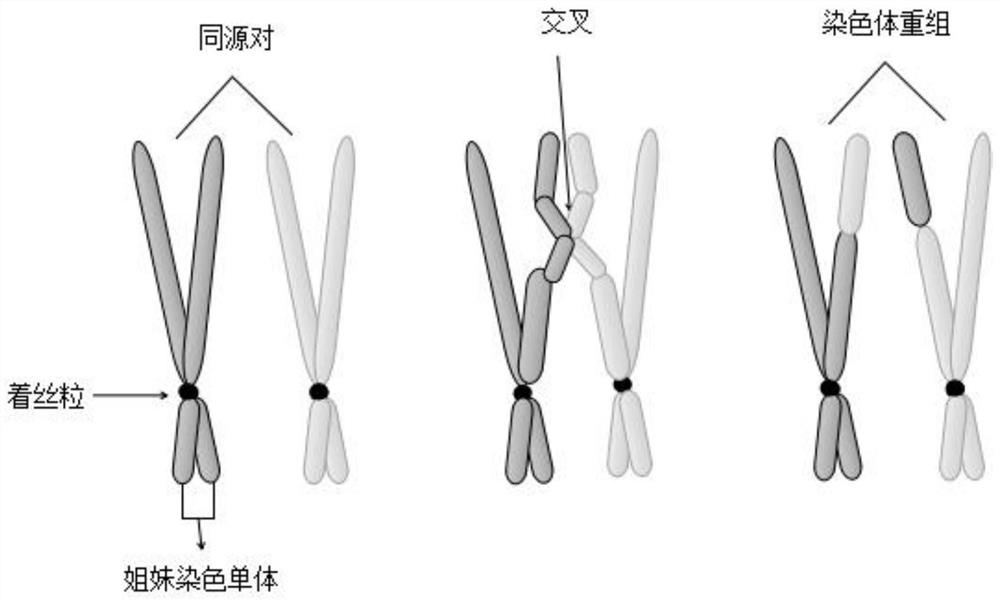
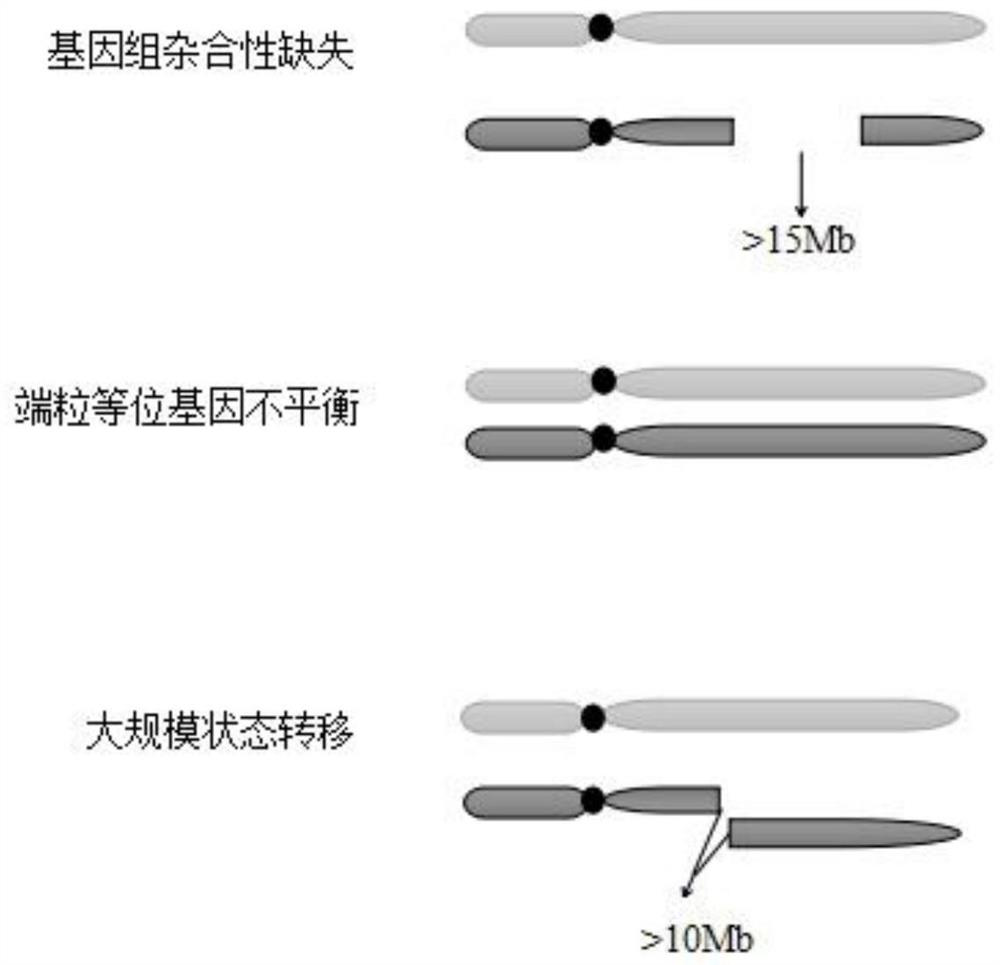
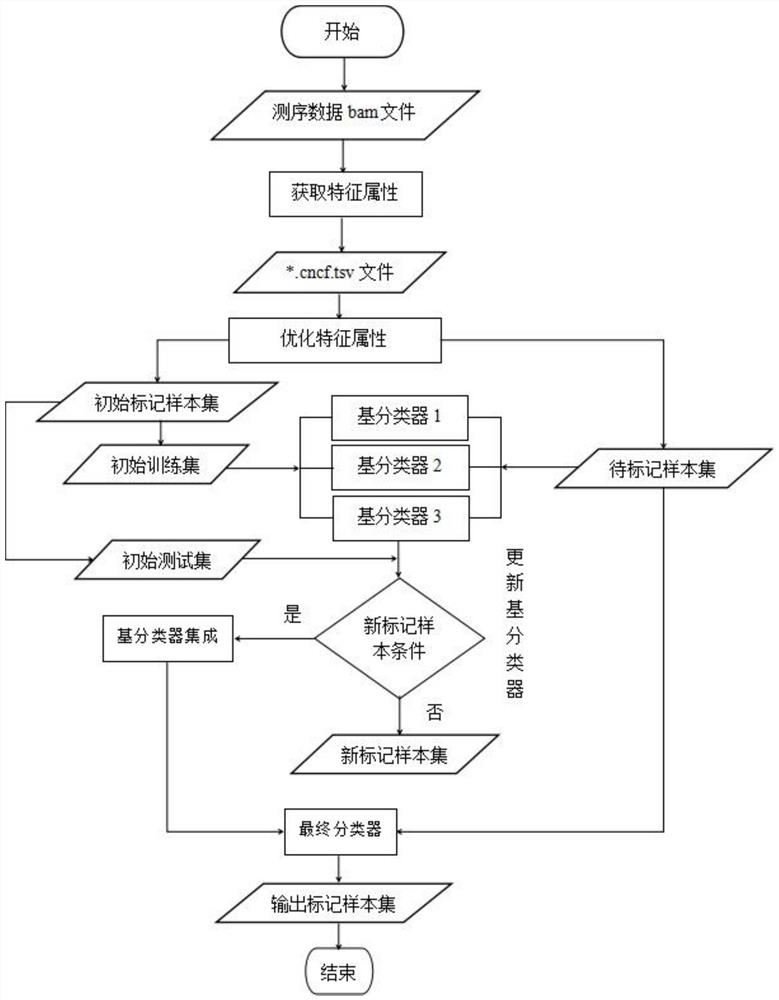
[0044] The invention provides a method for judging homologous recombination defects based on DNA sequencing data, which obtains characteristic attributes; extracts valid data; Three Base Classifiers H for Processing Efficiency 1 , H 2 , H 3 ; Pair H one by one 1 , H 2 , H 3 Carry out training to obtain an expanded training set, and iteratively update the base classifier; use the triple learning framework completed by training to mark the unlabeled sample set U, and complete the HRD status judgment according to the labeling result. The invention breaks through the limitation of using a small amount of local features such as genomic instability state for HRD determination, solves the difficulty of the extremely small number of samples with known HRD states, and realizes machine learning through multi-feature attributes when there is a small amount of data To improve the performance of HRD judgment method.
[0045] see image 3 , a method for determining homologous recombi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com