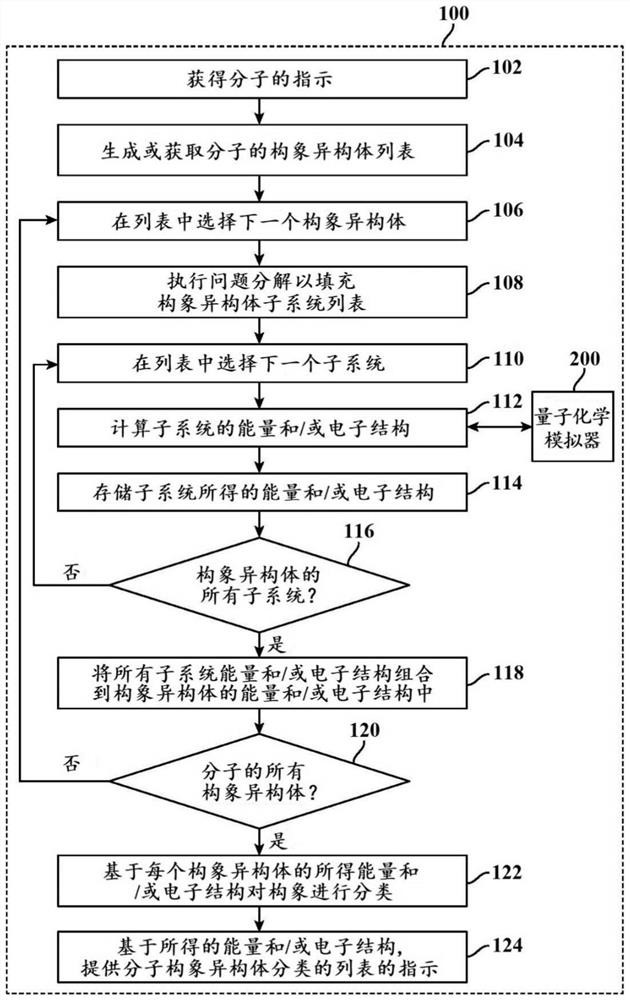
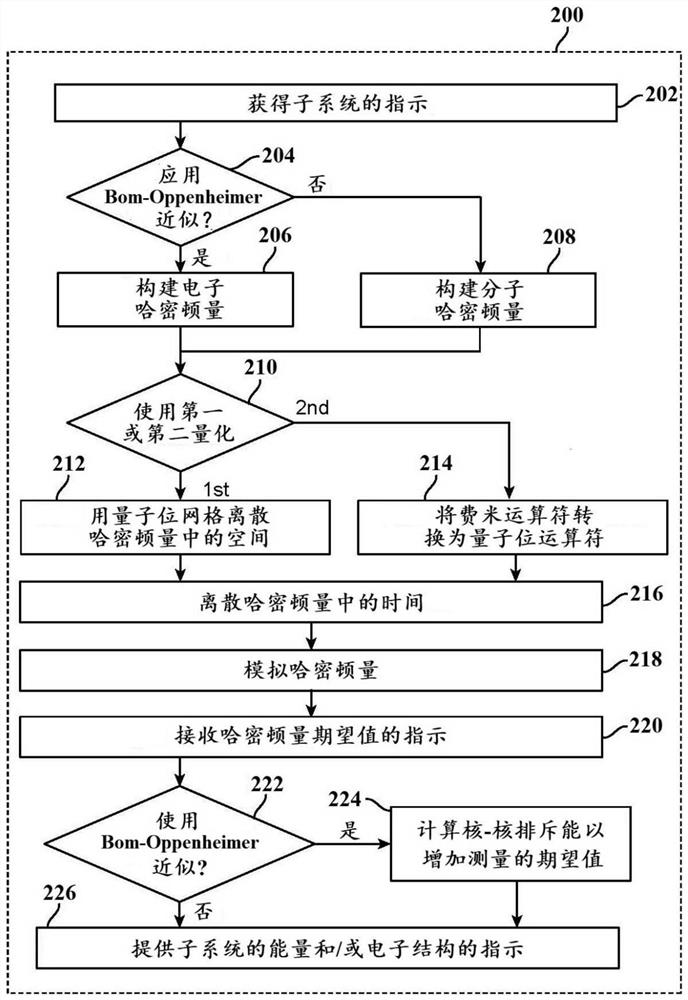
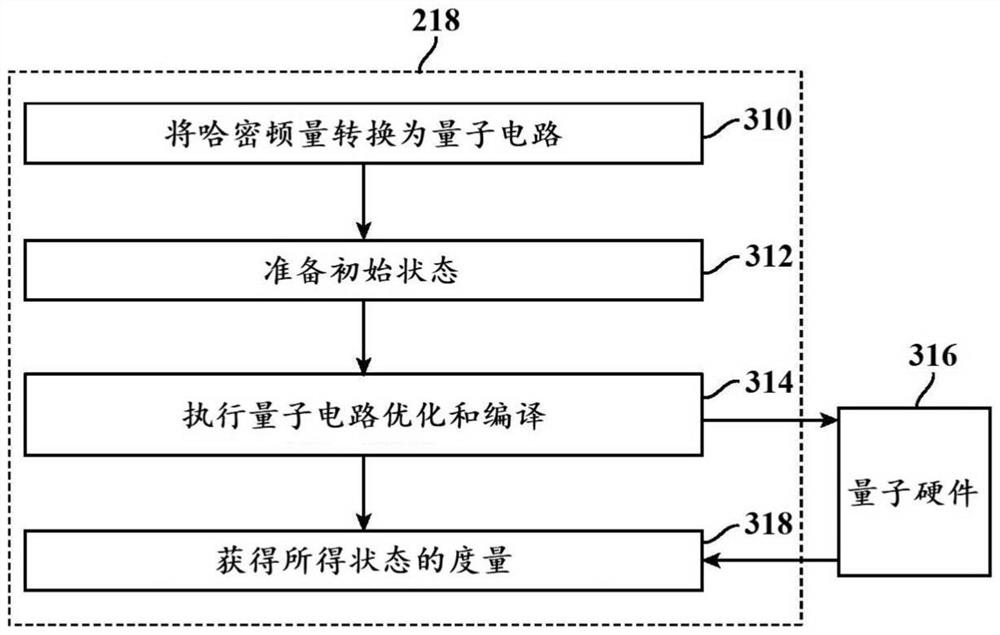
Methods and systems for quantum computing enabled molecular ab initio simulations using quantum-classical computing hardware
A quantum and molecular technology, applied in quantum computers, calculation models, calculations, etc., to achieve the effect of reducing the demand for resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0147] Example 1 (n-heptane)
[0148] The correlation between the total quantum mechanical energy calculations with and without PD was investigated for different conformations of the compounds. The simulation results of the fixed conformation of PD may not be within the range of chemical accuracy. However, if this is due to a systematic error, comparing two erroneous results for different conformers of the same molecule removes the error and provides an accurate relative quantum mechanical energy difference between the two conformations of the molecule. Thus, even without the best precise estimate of the total quantum mechanical energy for each individual conformer, this method can be used to accurately select the best conformer based on their total quantum mechanical energy values ( For example, the most stable conformer). Under this approach, more aggressive PD techniques (e.g., DC with a relatively small buffer size) can be used to find the best conformer from the poo...
example 2
[0152] Example 2 (3-methylheptane)
[0153] As observed, both FMO and DC work relatively well for simple polymer systems. Next, if Figure 7 As shown, by moving a methyl group to the carbon atom at the "3" position of n-heptane to generate 3-methylheptane, a diverse energy spectrum was generated for detection. The introduction of a methyl group into the "3" position makes the molecule asymmetric. As in the case of n-heptane, the conformational set of 3-methylheptane was generated by changing the four dihedral angles by 120 degrees (trans, side, side') and removing the high-energy conformations to obtain 65 conformations .
[0154] Figure 8 The quantum mechanical energy distribution (energy relative to the lowest energy) obtained by CCSD is shown to illustrate how one methyl group modulates and diversifies the quantum mechanical energy spectrum from n-heptane. Figure 9 A comparison between the exact CCSD and DC-CCSD results and between the exact CCSD and FMO-CCSD resul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com