Genome of low porcine backfat thickness and molecular marking method and breeding application
A molecular marker, pig backfat thick technology, applied in the field of molecular genetics and breeding, can solve the problems of increasing breeding costs, slow genetic progress, and lengthening the generation interval.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The specific details in the description of the present invention are only for fully understanding the embodiments of the present invention, but those skilled in the art should know that the implementation of the present invention is not limited to these details. In addition, well-known structures and functions are not described or shown in detail to avoid obscuring the gist of the embodiments of the present invention. Those of ordinary skill in the art can understand the specific meanings of the above terms in the present invention in specific situations.
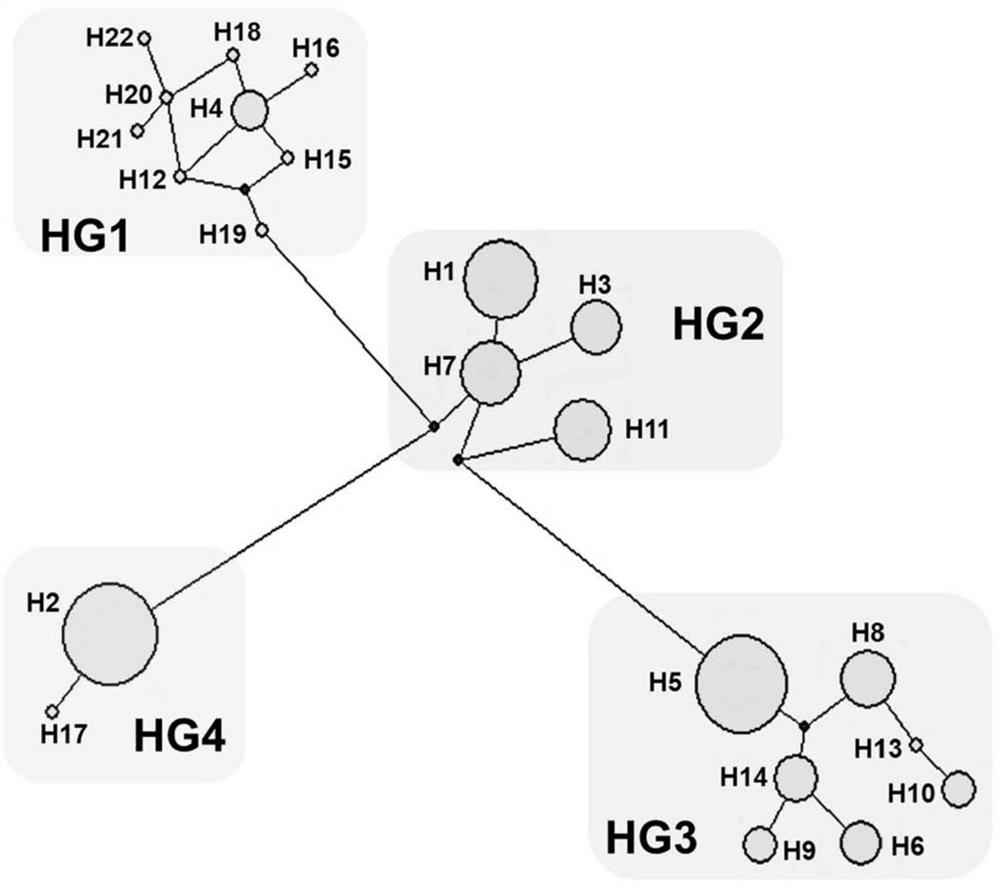
[0028] A specific embodiment of the present invention: pig low backfat genome, characterized in that the nucleotide sequence of the genome is the sequence shown in Seq ID No: 4, which is related to the low backfat trait of pigs.
[0029] The gene sequence Seq ID No: 4 is the nucleotide sequence of the haplotype H4 existing in the D-Loop region of the pig mitochondrial genome.
[0030] In the genome, the sequence sho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com