A method for mapping macromolecular complex structures to genome and mutation databases
A compound and genome technology, applied in the field of structure and genome information research, can solve the problems affecting the understanding of the molecular mechanism of pathogenic mutations, without considering RNA/DNA mutations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
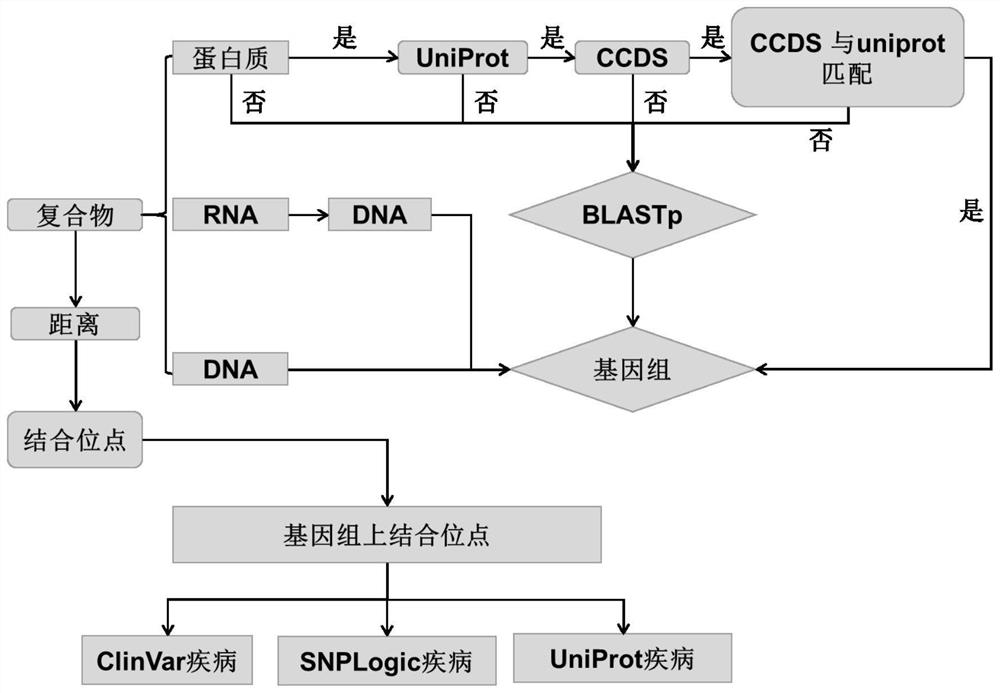
Image
Examples
Embodiment
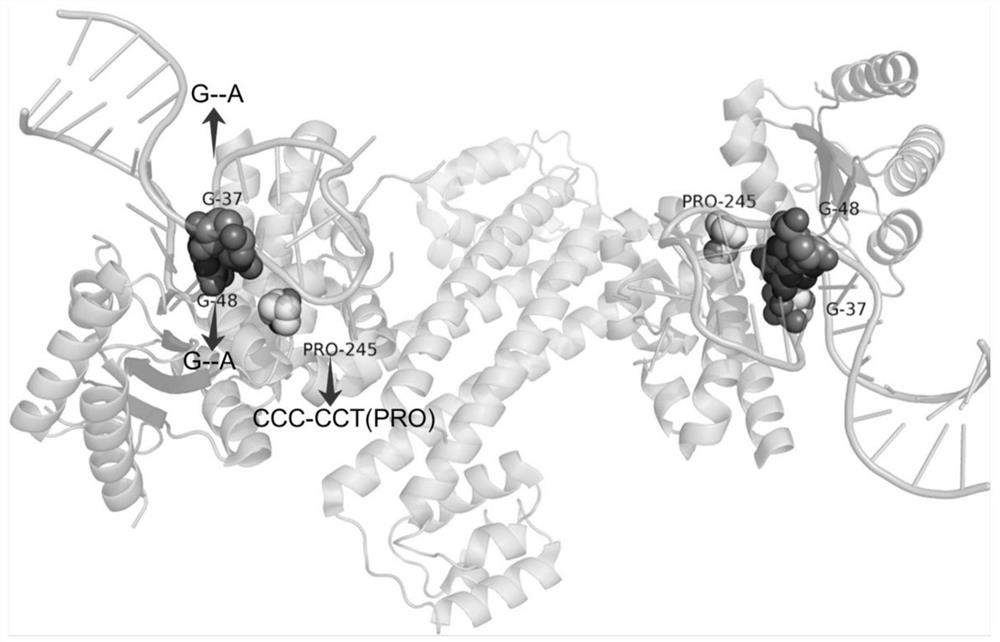
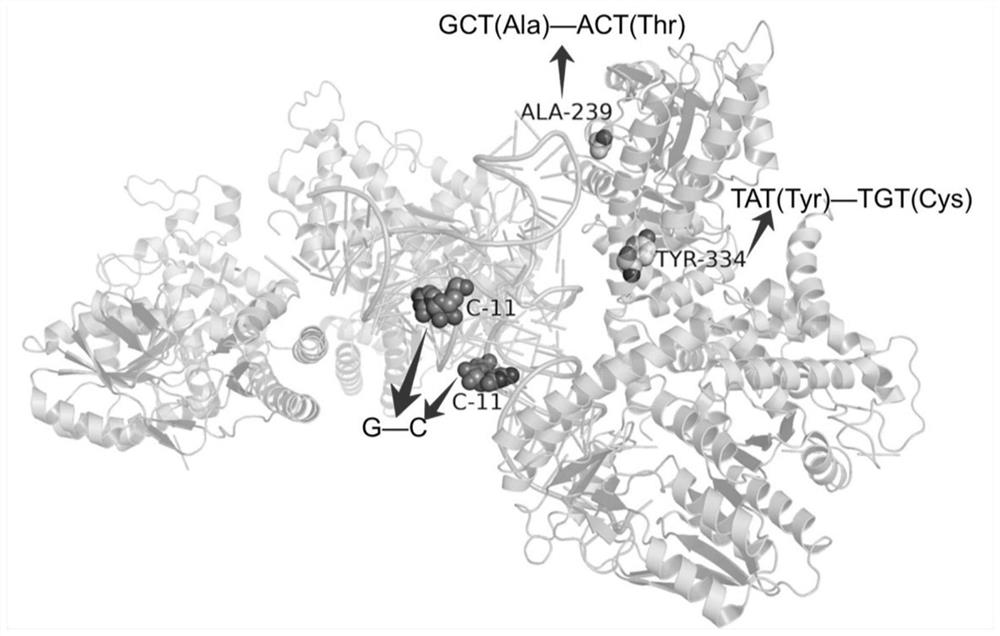
[0097] After the above steps, the invention found that a large number of SNPs occurred at the protein-RNA / DNA / protein interaction interface, as shown in Table 1:
[0098] Table 1: Mapping of protein-macromolecular complex structures to different disease databases
[0099]
[0100] The number before " / " indicates the number of chains with mutations on the interaction interface; the number after " / " indicates the number of chains with mutations, and "-" indicates that no chains could be mapped to the database.
[0101] Although it is possible to use sequencing or previous studies to understand which gene has information about the disease caused by the mutation, it does not tell us the structure of the functional three-dimensional complex corresponding to that gene, which can sometimes be detrimental to our understanding of the disease-causing mechanism. reason. However, the present invention can map the complex structure to the genome and explore how associated mutations (SN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com