Genome copy number variation detection method and device
A copy number variation and genome technology, applied in the fields of genomics, proteomics, instruments, etc., can solve the problems that need to be improved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
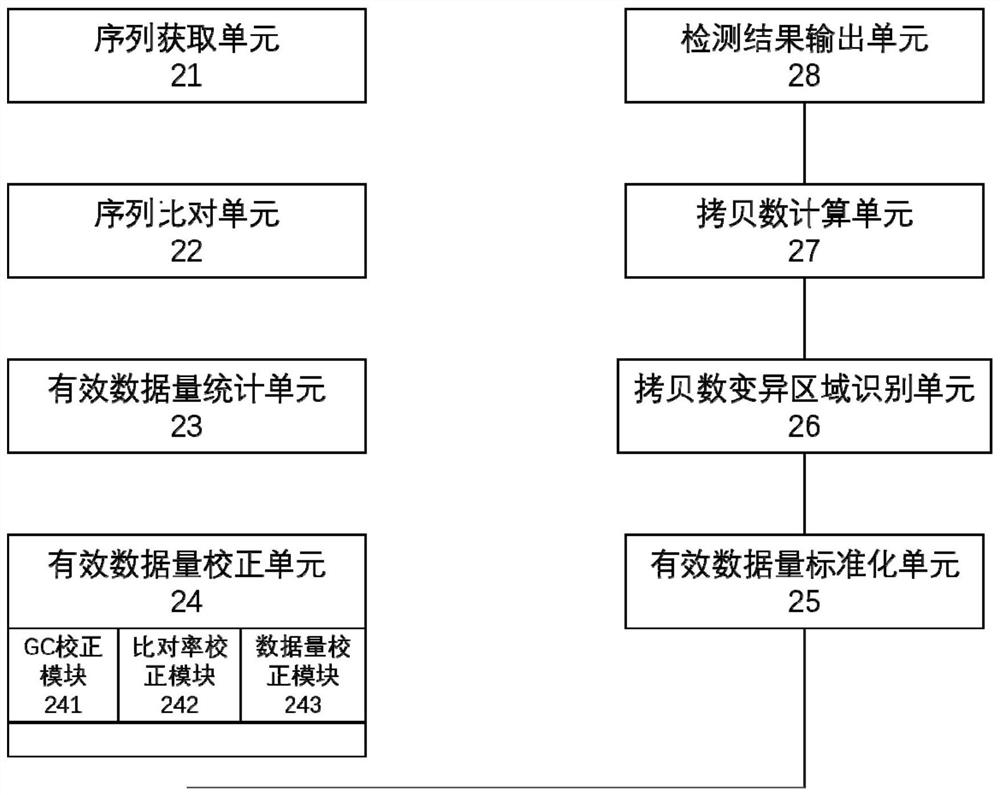
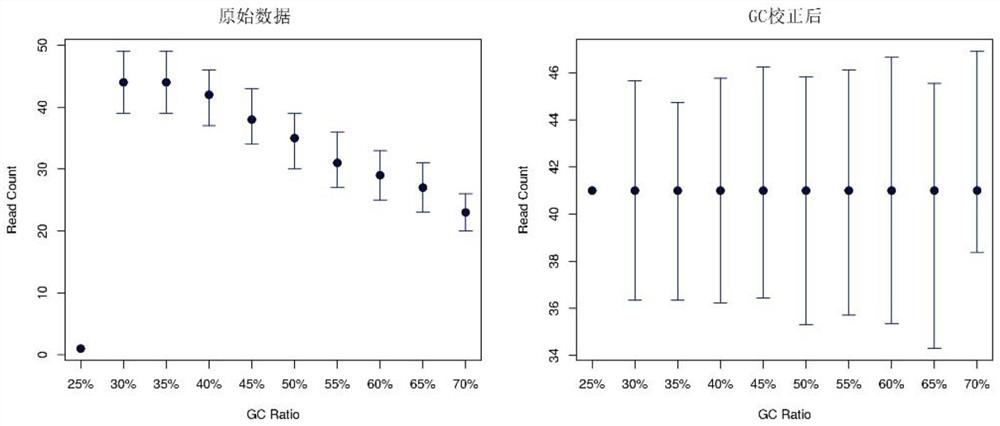
Image
Examples
Embodiment 1
[0065] Example 1. Detection of genome copy number variation according to the method of the present invention
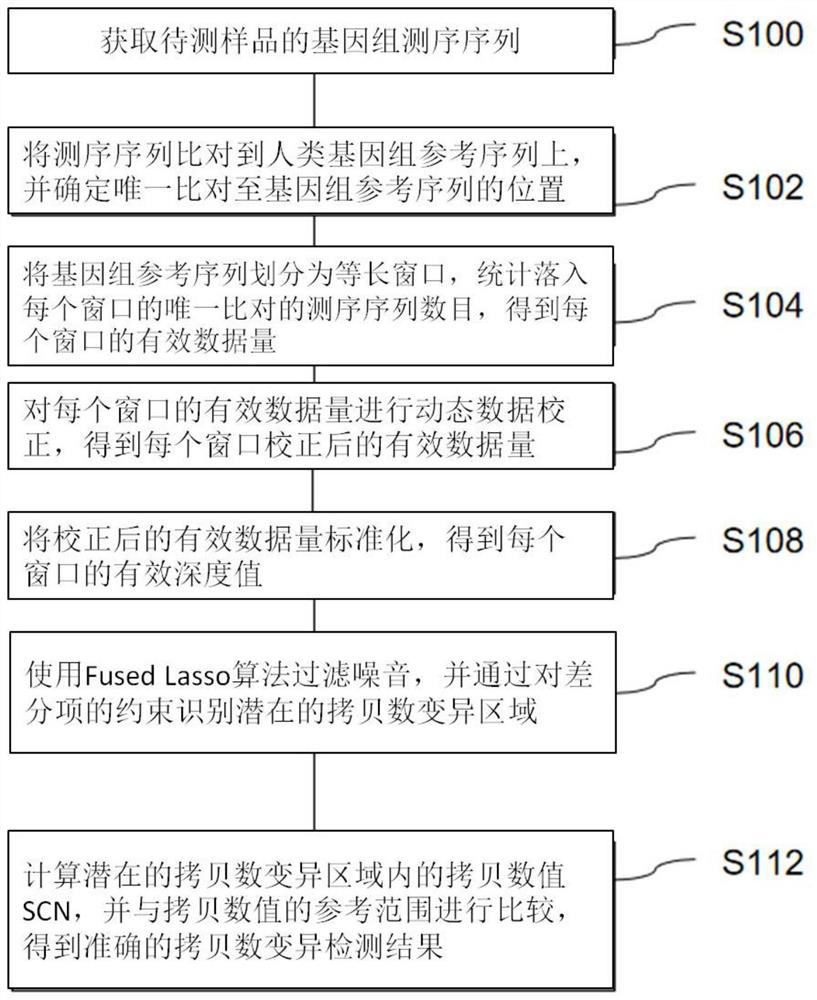
[0066] Detecting whether there is copy number variation in the sample to be tested according to the method of the present invention specifically includes the following steps (see figure 1 ):
[0067] S100: Obtain the genome sequencing sequence of the sample to be tested.
[0068] Obtain high-throughput sequencing data of 9 flow-through product samples. The data were constructed by using the Chromosomal Copy Number Variation Detection Kit (Reversible Terminal Termination Sequencing) for library construction, and using the NextSeq CN500 Gene Sequencer (Medical Device Registration Number: National Machinery Note 20153400460, Hangzhou Berry Hekang Gene Diagnostic Technology Co., Ltd. Ltd.) obtained.
[0069] S102: Align the sequencing sequence to the human genome reference sequence, and determine the unique alignment to the genome reference sequence position.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com