An ssr marker ch0211 for identifying soybean resistance to soybean mosaic virus sc3 strain and its detection method and application
A technology for soybean mosaic virus and detection method, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. The effect of improving the efficiency of disease resistance breeding, high economic benefits and saving manpower
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The experimental materials are:
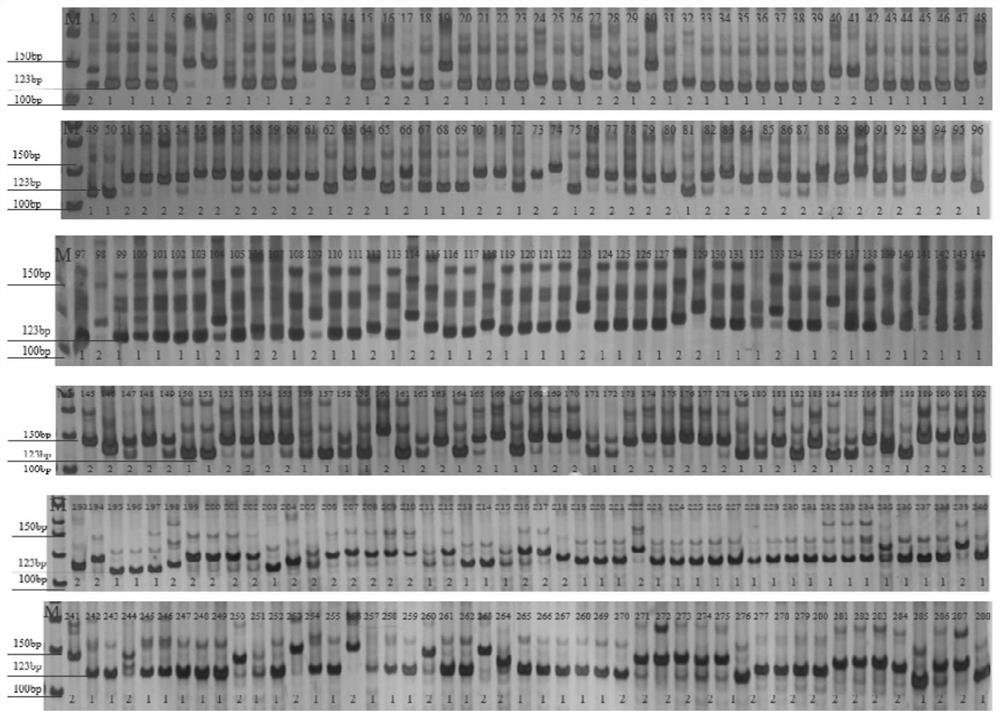
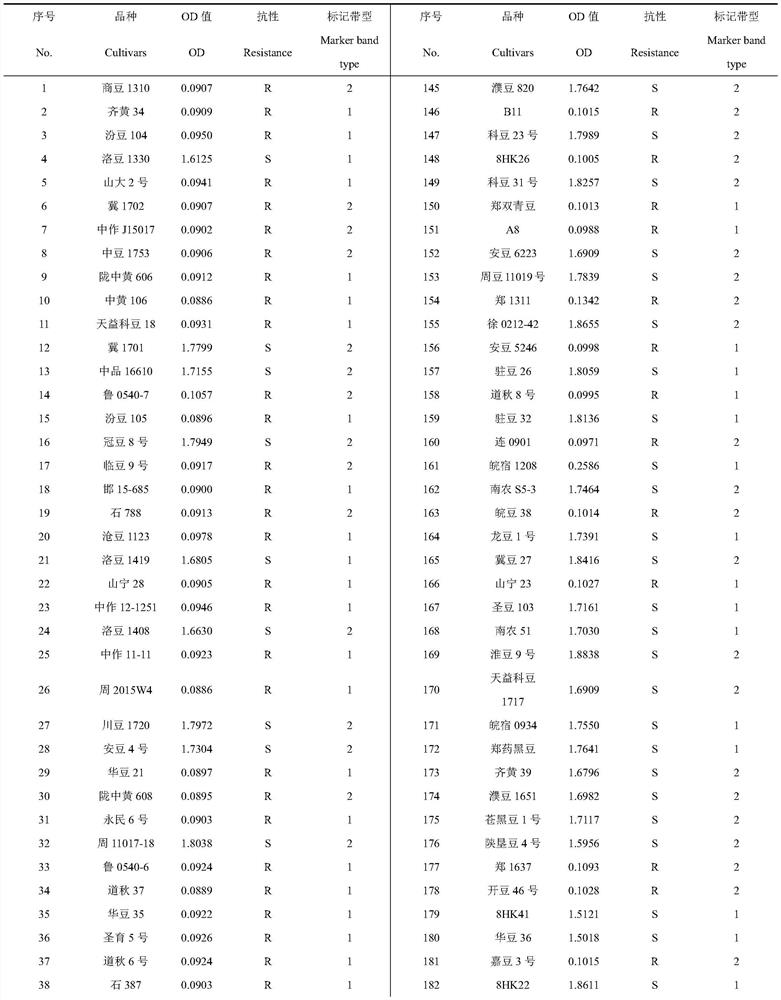
[0037] There are 288 varieties newly bred by major domestic breeding units in 2018 in the National Soybean Improvement Center of Nanjing Agricultural University.
[0038] The sequence of the SSR marker CH0211 primer used is:
[0039] Upstream primer: 5'-CAGGTCGATAAGTCTATGT-3' (SEQ ID NO.1);
[0040] Downstream primer: 5'-ATATGAACTCGGTCTGTTG-3' (SEQ ID NO. 2).
[0041] The above-marked primers were used to detect the resistance of soybean varieties to soybean mosaic virus SC3 strain.
[0042] S1. Among the 288 varieties newly bred by major domestic breeding units in 2018 stored in the National Soybean Improvement Center of Nanjing Agricultural University (Table 1), the genomic DNA was extracted according to the operation guide of the Plant Genomic DNA Extraction Kit (Kangwei, CW0531M), and the The SSR-labeled CH0211 was amplified by PCR, and the PCR amplification system was 10 μL, including 5 μL of 2×Taq PlusMasterMix, 0.5 μL of the ups...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com